NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors
- PMID: 27782878
- PMCID: PMC5111884
- DOI: 10.7554/eLife.18790
NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors
Abstract
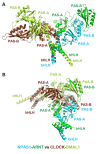
The neuronal PAS domain proteins NPAS1 and NPAS3 are members of the basic helix-loop-helix-PER-ARNT-SIM (bHLH-PAS) family, and their genetic deficiencies are linked to a variety of human psychiatric disorders including schizophrenia, autism spectrum disorders and bipolar disease. NPAS1 and NPAS3 must each heterodimerize with the aryl hydrocarbon receptor nuclear translocator (ARNT), to form functional transcription complexes capable of DNA binding and gene regulation. Here we examined the crystal structures of multi-domain NPAS1-ARNT and NPAS3-ARNT-DNA complexes, discovering each to contain four putative ligand-binding pockets. Through expanded architectural comparisons between these complexes and HIF-1α-ARNT, HIF-2α-ARNT and CLOCK-BMAL1, we show the wider mammalian bHLH-PAS family is capable of multi-ligand-binding and presents as an ideal class of transcription factors for direct targeting by small-molecule drugs.
Keywords: Hypoxia-Inducible Factor; Neuronal PAS Proteins; PAS domains; bHLH-PAS proteins; biophysics; crystal structure; drug discovery; human; ligand binding pockets; mouse; psychiatric disorders; structural biology; transcription factor.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures






References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research. 2014;42:W252–W258. doi: 10.1093/nar/gku340. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

