Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex
- PMID: 27782884
- PMCID: PMC5153249
- DOI: 10.7554/eLife.20934
Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex
Abstract
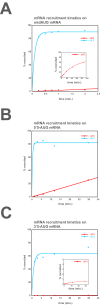
Eukaryotic translation initiation factor 3 (eIF3) is a central player in recruitment of the pre-initiation complex (PIC) to mRNA. We probed the effects on mRNA recruitment of a library of S. cerevisiae eIF3 functional variants spanning its 5 essential subunits using an in vitro-reconstituted system. Mutations throughout eIF3 disrupt its interaction with the PIC and diminish its ability to accelerate recruitment to a native yeast mRNA. Alterations to the eIF3a CTD and eIF3b/i/g significantly slow mRNA recruitment, and mutations within eIF3b/i/g destabilize eIF2•GTP•Met-tRNAi binding to the PIC. Using model mRNAs lacking contacts with the 40S entry or exit channels, we uncovered a critical role for eIF3 requiring the eIF3a NTD, in stabilizing mRNA interactions at the exit channel, and an ancillary role at the entry channel requiring residues of the eIF3a CTD. These functions are redundant: defects at each channel can be rescued by filling the other channel with mRNA.
Keywords: S. cerevisiae; biochemistry; biophysics; eIF3; initiation; mRNA recruitment; ribosome; structural biology; translation; yeast.
Conflict of interest statement
AGH: Reviewing editor, eLife. The other authors declare that no competing interests exist.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

