γ-Protocadherin structural diversity and functional implications
- PMID: 27782885
- PMCID: PMC5106212
- DOI: 10.7554/eLife.20930
γ-Protocadherin structural diversity and functional implications
Abstract
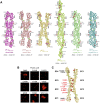
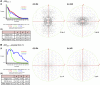
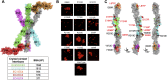
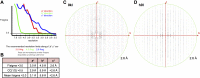
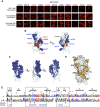
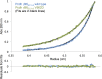
Stochastic cell-surface expression of α-, β-, and γ-clustered protocadherins (Pcdhs) provides vertebrate neurons with single-cell identities that underlie neuronal self-recognition. Here we report crystal structures of ectodomain fragments comprising cell-cell recognition regions of mouse γ-Pcdhs γA1, γA8, γB2, and γB7 revealing trans-homodimers, and of C-terminal ectodomain fragments from γ-Pcdhs γA4 and γB2, which depict cis-interacting regions in monomeric form. Together these structures span the entire γ-Pcdh ectodomain. The trans-dimer structures reveal determinants of γ-Pcdh isoform-specific homophilic recognition. We identified and structurally mapped cis-dimerization mutations to the C-terminal ectodomain structures. Biophysical studies showed that Pcdh ectodomains from γB-subfamily isoforms formed cis dimers, whereas γA isoforms did not, but both γA and γB isoforms could interact in cis with α-Pcdhs. Together, these data show how interaction specificity is distributed over all domains of the γ-Pcdh trans interface, and suggest that subfamily- or isoform-specific cis-interactions may play a role in the Pcdh-mediated neuronal self-recognition code.
Keywords: biophysics; clustered protocadherins; crystal structure; mouse; neuronal self-avoidance; neuroscience; protein diversity; protein-protein recognition; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
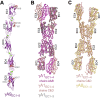












References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

