NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription
- PMID: 27783096
- PMCID: PMC5309293
- DOI: 10.1007/s00018-016-2398-4
NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription
Abstract
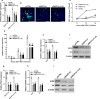
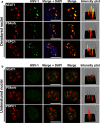
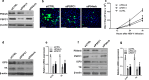
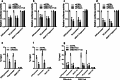
Nuclear paraspeckle assembly transcript 1 (NEAT1) is the crucial structural platform of paraspeckles, which is one type of nuclear bodies. As a stress-induced lncRNA, the expression of NEAT1 increases in response to viral infection, but little is known about the role of NEAT1 or paraspeckles in the replication of herpes simplex virus-1 (HSV-1). Here, we demonstrate that HSV-1 infection increases NEAT1 expression and paraspeckle formation in a STAT3-dependent manner. NEAT1 and other paraspeckle protein components, P54nrb and PSPC1, can associate with HSV-1 genomic DNA. By binding with STAT3, PSPC1 is required for the recruitment of STAT3 to paraspeckles and facilitates the interaction between STAT3 and viral gene promoters, finally increasing viral gene expression and viral replication. Furthermore, thermosensitive gel containing NEAT1 siRNA or STAT3 siRNA effectively healed the skin lesions caused by HSV-1 infection in mice. Our results provide insight into the roles of lncRNAs in the epigenetic control of viral genes and into the function of paraspeckles.
Keywords: Gene regulation; HSV-1; NEAT1; Paraspeckle; STAT3; Viral replication.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

