Go it alone: four-electron oxidations by mononuclear non-heme iron enzymes
- PMID: 27783267
- PMCID: PMC5352498
- DOI: 10.1007/s00775-016-1399-y
Go it alone: four-electron oxidations by mononuclear non-heme iron enzymes
Abstract
This review discusses the current mechanistic understanding of a group of mononuclear non-heme iron-dependent enzymes that catalyze four-electron oxidation of their organic substrates without the use of any cofactors or cosubstrates. One set of enzymes acts on α-ketoacid-containing substrates, coupling decarboxylation to oxygen activation. This group includes 4-hydroxyphenylpyruvate dioxygenase, 4-hydroxymandelate synthase, and CloR involved in clorobiocin biosynthesis. A second set of enzymes acts on substrates containing a thiol group that coordinates to the iron. This group is comprised of isopenicillin N synthase, thiol dioxygenases, and enzymes involved in the biosynthesis of ergothioneine and ovothiol. The final group of enzymes includes HEPD and MPnS that both carry out the oxidative cleavage of the carbon-carbon bond of 2-hydroxyethylphosphonate but generate different products. Commonalities amongst many of these enzymes are discussed and include the initial substrate oxidation by a ferric-superoxo-intermediate and a second oxidation by a ferryl species.
Keywords: Ferryl; Iron oxo; Non-heme iron; Oxidase; Oxygenase; Superoxo.
Figures

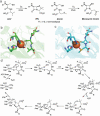

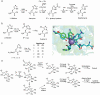
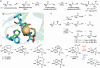
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

