Middle East Respiratory Coronavirus Accessory Protein 4a Inhibits PKR-Mediated Antiviral Stress Responses
- PMID: 27783669
- PMCID: PMC5081173
- DOI: 10.1371/journal.ppat.1005982
Middle East Respiratory Coronavirus Accessory Protein 4a Inhibits PKR-Mediated Antiviral Stress Responses
Abstract
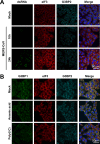
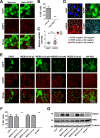
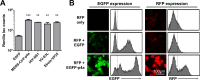
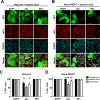
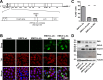
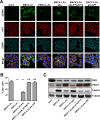
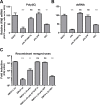
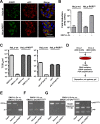
Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe respiratory infections that can be life-threatening. To establish an infection and spread, MERS-CoV, like most other viruses, must navigate through an intricate network of antiviral host responses. Besides the well-known type I interferon (IFN-α/β) response, the protein kinase R (PKR)-mediated stress response is being recognized as an important innate response pathway. Upon detecting viral dsRNA, PKR phosphorylates eIF2α, leading to the inhibition of cellular and viral translation and the formation of stress granules (SGs), which are increasingly recognized as platforms for antiviral signaling pathways. It is unknown whether cellular infection by MERS-CoV activates the stress response pathway or whether the virus has evolved strategies to suppress this infection-limiting pathway. Here, we show that cellular infection with MERS-CoV does not lead to the formation of SGs. By transiently expressing the MERS-CoV accessory proteins individually, we identified a role of protein 4a (p4a) in preventing activation of the stress response pathway. Expression of MERS-CoV p4a impeded dsRNA-mediated PKR activation, thereby rescuing translation inhibition and preventing SG formation. In contrast, p4a failed to suppress stress response pathway activation that is independent of PKR and dsRNA. MERS-CoV p4a is a dsRNA binding protein. Mutation of the dsRNA binding motif in p4a disrupted its PKR antagonistic activity. By inserting p4a in a picornavirus lacking its natural PKR antagonist, we showed that p4a exerts PKR antagonistic activity also under infection conditions. However, a recombinant MERS-CoV deficient in p4a expression still suppressed SG formation, indicating the expression of at least one other stress response antagonist. This virus also suppressed the dsRNA-independent stress response pathway. Thus, MERS-CoV interferes with antiviral stress responses using at least two different mechanisms, with p4a suppressing the PKR-dependent stress response pathway, probably by sequestering dsRNA. MERS-CoV p4a represents the first coronavirus stress response antagonist described.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Inhibition of Stress Granule Formation by Middle East Respiratory Syndrome Coronavirus 4a Accessory Protein Facilitates Viral Translation, Leading to Efficient Virus Replication.J Virol. 2018 Sep 26;92(20):e00902-18. doi: 10.1128/JVI.00902-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30068649 Free PMC article.
-
Antagonism of dsRNA-Induced Innate Immune Pathways by NS4a and NS4b Accessory Proteins during MERS Coronavirus Infection.mBio. 2019 Mar 26;10(2):e00319-19. doi: 10.1128/mBio.00319-19. mBio. 2019. PMID: 30914508 Free PMC article.
-
Middle East Respiratory Syndrome Coronavirus NS4b Protein Inhibits Host RNase L Activation.mBio. 2016 Mar 29;7(2):e00258. doi: 10.1128/mBio.00258-16. mBio. 2016. PMID: 27025250 Free PMC article.
-
Modulation of the immune response by Middle East respiratory syndrome coronavirus.J Cell Physiol. 2019 Mar;234(3):2143-2151. doi: 10.1002/jcp.27155. Epub 2018 Aug 26. J Cell Physiol. 2019. PMID: 30146782 Free PMC article. Review.
-
Severe acute respiratory syndrome vs. the Middle East respiratory syndrome.Curr Opin Pulm Med. 2014 May;20(3):233-41. doi: 10.1097/MCP.0000000000000046. Curr Opin Pulm Med. 2014. PMID: 24626235 Review.
Cited by
-
Proteolytic Activities of Enterovirus 2A Do Not Depend on Its Interaction with SETD3.Viruses. 2022 Jun 22;14(7):1360. doi: 10.3390/v14071360. Viruses. 2022. PMID: 35891342 Free PMC article.
-
Glycogen synthase kinase (GSK)-3 and the double-strand RNA-dependent kinase, PKR: When two kinases for the common good turn bad.Biochim Biophys Acta Mol Cell Res. 2020 Oct;1867(10):118769. doi: 10.1016/j.bbamcr.2020.118769. Epub 2020 Jun 5. Biochim Biophys Acta Mol Cell Res. 2020. PMID: 32512016 Free PMC article. Review.
-
Functions of Coronavirus Accessory Proteins: Overview of the State of the Art.Viruses. 2021 Jun 13;13(6):1139. doi: 10.3390/v13061139. Viruses. 2021. PMID: 34199223 Free PMC article. Review.
-
Transgene expression in the genome of Middle East respiratory syndrome coronavirus based on a novel reverse genetics system utilizing Red-mediated recombination cloning.J Gen Virol. 2017 Oct;98(10):2461-2469. doi: 10.1099/jgv.0.000919. J Gen Virol. 2017. PMID: 28984231 Free PMC article.
-
Translational control of coronaviruses.Nucleic Acids Res. 2020 Dec 16;48(22):12502-12522. doi: 10.1093/nar/gkaa1116. Nucleic Acids Res. 2020. PMID: 33264393 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

