Bacillus subtilis 5'-nucleotidases with various functions and substrate specificities
- PMID: 27784292
- PMCID: PMC5080769
- DOI: 10.1186/s12866-016-0866-5
Bacillus subtilis 5'-nucleotidases with various functions and substrate specificities
Abstract
Background: In Escherichia coli, nagD, yrfG, yjjG, yieH, yigL, surE, and yfbR encode 5'-nucleotidases that hydrolyze the phosphate group of 5'-nucleotides. In Bacillus subtilis, genes encoding 5'-nucleotidase have remained to be identified.
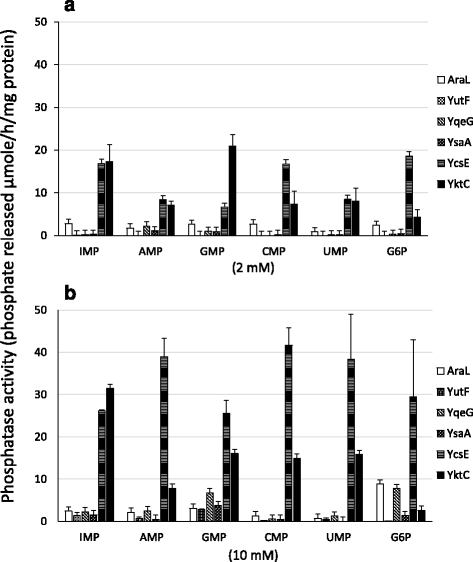
Results: We found that B. subtilis ycsE, araL, yutF, ysaA, and yqeG show suggestive similarities to nagD. Here, we expressed them in E. coli to purify the respective His6-tagged proteins. YcsE exhibited significant 5'-nucleotidase activity with a broader specificity, whereas the other four enzymes had rather weak but suggestive activities with various capacities and substrate specificities. In contrast, B. subtilis yktC shares high similarity with E. coli suhB encoding an inositol monophosphatase. YktC exhibited inositol monophosphatase activity as well as 5'-nucleotidase activity preferential for GMP and IMP. The ycsE, yktC, and yqeG genes are induced by oxidative stress and were dispensable, although yqeG was required to maintain normal growth on solid medium. In the presence of diamide, only mutants lacking yktC exhibited enhanced growth defects, whereas the other mutants without ycsE or yqeG did not.
Conclusions: Accordingly, in B. subtilis, at least YcsE and YktC acted as major 5'-nucleotidases and the four minor enzymes might function when the intracellular concentrations of substrates are sufficiently high. In addition, YktC is involved in resistance to oxidative stress caused by diamide, while YqeG is necessary for normal colony formation on solid medium.
Keywords: 5′-nucleotidase; Bacillus subtilis; Haloacid dehalogenase superfamily; Inositol monophosphatase; Inositol phosphate; Nucleoside/nucleotide metabolism; Oxidative stress; Phosphatase; Protein motif.
Figures


References
-
- Tamao Y, Noguchi K, Sakai-Tomita Y, Hama H, Shimamoto T, Kanazawa H, Tsuda M, Tsuchiya T. Sequence analysis of nutA gene encoding membrane-bound Cl(−)-dependent 5′-nucleotidase of Vibrio parahaemolyticus. J Biochem. 1991;109:24–29. - PubMed
-
- Burns DM, Beacham IR. Nucleotide sequence and transcriptional analysis of the E. coli ushA gene, encoding periplasmic UDP-sugar hydrolase (5′-nucleotidase): regulation of the ushA gene, and the signal sequence of its encoded protein product. Nucleic Acids Res. 1986;14:4325–4342. doi: 10.1093/nar/14.10.4325. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

