Integrating cancer genomic data into electronic health records
- PMID: 27784327
- PMCID: PMC5081968
- DOI: 10.1186/s13073-016-0371-3
Integrating cancer genomic data into electronic health records
Abstract
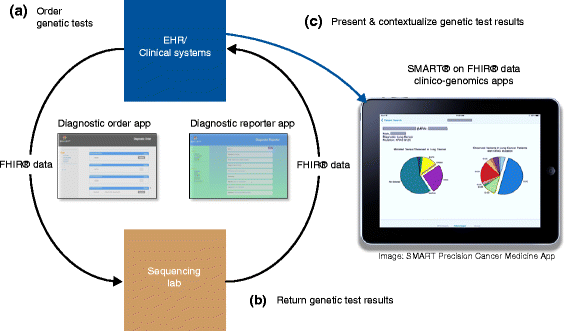
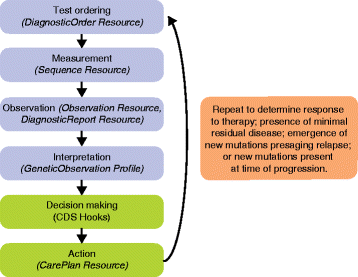
The rise of genomically targeted therapies and immunotherapy has revolutionized the practice of oncology in the last 10-15 years. At the same time, new technologies and the electronic health record (EHR) in particular have permeated the oncology clinic. Initially designed as billing and clinical documentation systems, EHR systems have not anticipated the complexity and variety of genomic information that needs to be reviewed, interpreted, and acted upon on a daily basis. Improved integration of cancer genomic data with EHR systems will help guide clinician decision making, support secondary uses, and ultimately improve patient care within oncology clinics. Some of the key factors relating to the challenge of integrating cancer genomic data into EHRs include: the bioinformatics pipelines that translate raw genomic data into meaningful, actionable results; the role of human curation in the interpretation of variant calls; and the need for consistent standards with regard to genomic and clinical data. Several emerging paradigms for integration are discussed in this review, including: non-standardized efforts between individual institutions and genomic testing laboratories; "middleware" products that portray genomic information, albeit outside of the clinical workflow; and application programming interfaces that have the potential to work within clinical workflow. The critical need for clinical-genomic knowledge bases, which can be independent or integrated into the aforementioned solutions, is also discussed.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

