Epitope-dependent Functional Effects of Celiac Disease Autoantibodies on Transglutaminase 2
- PMID: 27784785
- PMCID: PMC5207253
- DOI: 10.1074/jbc.M116.738161
Epitope-dependent Functional Effects of Celiac Disease Autoantibodies on Transglutaminase 2
Abstract
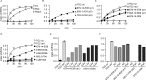
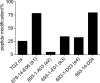
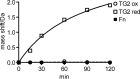
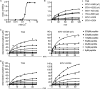
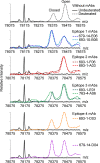
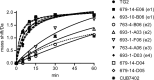
Transglutaminase 2 (TG2) is a Ca2+-dependent cross-linking enzyme involved in the pathogenesis of CD. We have previously characterized a panel of anti-TG2 mAbs generated from gut plasma cells of celiac patients and identified four epitopes (epitopes 1-4) located in the N-terminal part of TG2. Binding of the mAbs induced allosteric changes in TG2. Thus, we aimed to determine whether these mAbs could influence enzymatic activity through modulation of TG2 susceptibility to oxidative inactivation and Ca2+ affinity. All tested epitope 1 mAbs, as well as 679-14-D04, which recognizes a previously uncharacterized epitope, prevented oxidative inactivation and increased Ca2+ sensitivity of TG2. We have identified crucial residues for binding of 679-14-D04 located within a Ca2+ binding site. Epitope 1 mAbs and 679-14-D04, although recognizing separate epitopes, behaved similarly when assessing their effect on TG2 conformation, suggesting that the shared effects on TG2 function can be explained by induction of the same conformational changes. None of the mAbs targeting other epitopes showed these effects, but epitope 2 mAbs reduced the rate of TG2-catalyzed reactions. Collectively, these effects could be relevant to the pathogenesis of CD. In A20 B cells transduced with TG2-specific B-cell receptor, epitope 2-expressing cells had poorer uptake of TG2-gluten complexes and were less efficient in gluten epitope presentation to T cells than cells expressing an epitope 1 receptor. Thus, the ability of epitope 1-targeting B cells to keep TG2 active and protected from oxidation might explain why generation of epitope 1-targeting plasma cells seems to be favored in celiac patients.
Keywords: allosteric regulation; antibody; autoimmunity; capillary electrophoresis; celiac disease; enzyme catalysis; hydrogen/deuterium exchange; mass spectrometry (MS); transglutaminase.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Folk J. E. (1983) Mechanism and basis for specificity of transglutaminase-catalyzed ϵ-(γ-glutamyl) lysine bond formation. Adv. Enzymol. Relat. Areas Mol. Biol. 54, 1–56 - PubMed
-
- Folk J. E., Mullooly J. P., and Cole P. W. (1967) Mechanism of action of guinea pig liver transglutaminase: II. the role of metal in enzyme activation. J. Biol. Chem. 242, 1838–1844 - PubMed
-
- Kim S. Y. (2006) Transglutaminase 2 in inflammation. Front. Biosci. 11, 3026–3035 - PubMed
-
- Lorand L., and Graham R. M. (2003) Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4, 140–156 - PubMed
-
- Fesus L., and Piacentini M. (2002) Transglutaminase 2: an enigmatic enzyme with diverse functions. Trends Biochem. Sci. 27, 534–539 - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

