PrgU: a suppressor of sex pheromone toxicity in Enterococcus faecalis
- PMID: 27785854
- PMCID: PMC5263107
- DOI: 10.1111/mmi.13563
PrgU: a suppressor of sex pheromone toxicity in Enterococcus faecalis
Abstract
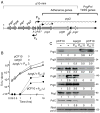
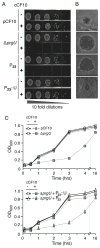
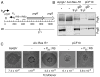
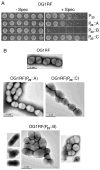
Upon sensing of the peptide pheromone cCF10, Enterococcus faecalis cells carrying pCF10 produce three surface adhesins (PrgA, PrgB or Aggregation Substance, PrgC) and the Prg/Pcf type IV secretion system and, in turn, conjugatively transfer the plasmid at high frequencies to recipient cells. Here, we report that cCF10 induction is highly toxic to cells sustaining a deletion of prgU, a small orf located immediately downstream of prgB on pCF10. Upon pheromone exposure, these cells overproduce the Prg adhesins and display impaired envelope integrity, as evidenced by antibiotic susceptibility, misplaced division septa and cell lysis. Compensatory mutations in regulatory loci controlling expression of pCF10-encoded prg/pcf genes, or constitutive PrgU overproduction, block production of the Prg adhesins and render cells insensitive to pheromone. Cells engineered to overproduce PrgB, even independently of other pCF10-encoded proteins, have severely compromised cell envelopes and strong growth defects. PrgU has an RNA-binding fold, and prgB-prgU gene pairs are widely distributed among E. faecalis isolates and other enterococcal and staphylococcal species. Together, our findings support a model in which PrgU proteins represent a novel class of RNA-binding regulators that act to mitigate toxicity accompanying overproduction of PrgB-like adhesins in E. faecalis and other clinically-important Gram-positive species.
© 2016 John Wiley & Sons Ltd.
Figures

Similar articles
-
Enterococcal PrgU Provides Additional Regulation of Pheromone-Inducible Conjugative Plasmids.mSphere. 2021 Jun 30;6(3):e0026421. doi: 10.1128/mSphere.00264-21. Epub 2021 Jun 9. mSphere. 2021. PMID: 34106752 Free PMC article.
-
Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence.Mol Microbiol. 2015 Feb;95(4):660-77. doi: 10.1111/mmi.12893. Epub 2014 Dec 30. Mol Microbiol. 2015. PMID: 25431047 Free PMC article.
-
Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10.J Mol Biol. 2000 Apr 7;297(4):861-75. doi: 10.1006/jmbi.2000.3628. J Mol Biol. 2000. PMID: 10736223
-
The sex pheromone system of Enterococcus faecalis. More than just a plasmid-collection mechanism?Eur J Biochem. 1994 Jun 1;222(2):235-46. doi: 10.1111/j.1432-1033.1994.tb18862.x. Eur J Biochem. 1994. PMID: 8020463 Review.
-
The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution.Philos Trans R Soc Lond B Biol Sci. 2007 Jul 29;362(1483):1185-93. doi: 10.1098/rstb.2007.2043. Philos Trans R Soc Lond B Biol Sci. 2007. PMID: 17360276 Free PMC article. Review.
Cited by
-
Single-Cell Analysis Reveals that the Enterococcal Sex Pheromone Response Results in Expression of Full-Length Conjugation Operon Transcripts in All Induced Cells.J Bacteriol. 2020 Mar 26;202(8):e00685-19. doi: 10.1128/JB.00685-19. Print 2020 Mar 26. J Bacteriol. 2020. PMID: 32041799 Free PMC article.
-
Suspended Materials in River Waters Differentially Enrich Class 1 Integron- and IncP-1 Plasmid-Carrying Bacteria in Sediments.Front Microbiol. 2018 Jul 2;9:1443. doi: 10.3389/fmicb.2018.01443. eCollection 2018. Front Microbiol. 2018. PMID: 30013540 Free PMC article.
-
Regulation of Gram-Positive Conjugation.Front Microbiol. 2019 May 22;10:1134. doi: 10.3389/fmicb.2019.01134. eCollection 2019. Front Microbiol. 2019. PMID: 31191478 Free PMC article. Review.
-
Enterococcal PrgU Provides Additional Regulation of Pheromone-Inducible Conjugative Plasmids.mSphere. 2021 Jun 30;6(3):e0026421. doi: 10.1128/mSphere.00264-21. Epub 2021 Jun 9. mSphere. 2021. PMID: 34106752 Free PMC article.
-
PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction.Mol Microbiol. 2018 Aug;109(3):291-305. doi: 10.1111/mmi.13980. Epub 2018 Jul 31. Mol Microbiol. 2018. PMID: 29723434 Free PMC article.
References
-
- Bae T, Kozlowicz BK, Dunny GM. Characterization of cis-acting prgQ mutants: evidence for two distinct repression mechanisms by Qa RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Mol Microbiol. 2004;51:271–281. - PubMed
-
- Bensing BA, Dunny GM. Pheromone-inducible expression of an aggregation protein in Enterococcus faecalis requires interaction of a plasmid-encoded RNA with components of the ribosome. Mol Microbiol. 1997;24:295–308. - PubMed
-
- Bensing BA, Manias DA, Dunny GM. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol Microbiol. 1997;24:285–294. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

