De novo methylation in male germ cells of the common marmoset monkey occurs during postnatal development and is maintained in vitro
- PMID: 27786608
- PMCID: PMC5687340
- DOI: 10.1080/15592294.2016.1248007
De novo methylation in male germ cells of the common marmoset monkey occurs during postnatal development and is maintained in vitro
Abstract
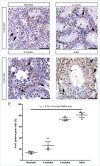
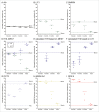
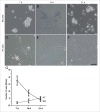
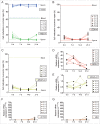
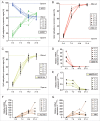
The timing of de novo DNA methylation in male germ cells during human testicular development is yet unsolved. Apart from that, the stability of established imprinting patterns in vitro is controversially discussed. This study aimed at determining the timing of DNA de novo methylation and at assessing the stability of the methylation status in vitro. We employed the marmoset monkey (Callithrix jacchus) as it is considered the best non-human primate model for human testicular development. We selected neonatal, pre-pubertal, pubertal, and adult animals (n = 3, each) and assessed germ cell global DNA methylation levels by 5-methyl cytosine staining, and Alu elements and gene-specific methylation (H19, LIT1, SNRPN, MEST, OCT4, MAGE-A4, and DDX-4) by pyrosequencing. De novo methylation is progressively established during postnatal primate development and continues until adulthood, a process that is different in most other species. Importantly, once established, methylation patterns remained stable, as demonstrated using in vitro cultures. Thus, the marmoset monkey is a unique model for the study of postnatal DNA methylation mechanisms in germ cells and for the identification of epimutations and their causes.
Keywords: de novo methylation; germ cell development; primate-specific DNA methylation patterns; spermatogonia.
Figures


References
-
- Monk D. Germline-derived DNA methylation and early embryo epigenetic reprogramming: The selected survival of imprints. Int J Biochem Cell Biol 2015; 67:128-138; PMID:25966912; https://doi.org/10.1016/j.biocel.2015.04.014 - DOI - PubMed
-
- Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. Reprogramming DNA methylation in the mammalian life cycle: Building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci 2013; 368:20110330; PMID:23166394; https://doi.org/10.1098/rstb.2011.0330 - DOI - PMC - PubMed
-
- Reik W, Walter J. Genomic imprinting: Parental influence on the genome. Nat Rev Genet 2001; 2:21-32; PMID:11253064; https://doi.org/10.1038/35047554 - DOI - PubMed
-
- Guibert S, Forne T, Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 2012; 22:633-41; PMID:22357612; https://doi.org/10.1101/gr.130997.111 - DOI - PMC - PubMed
-
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 2013; 339:448-52; PMID:23223451; https://doi.org/10.1126/science.1229277 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
