TCGA Expedition: A Data Acquisition and Management System for TCGA Data
- PMID: 27788220
- PMCID: PMC5082933
- DOI: 10.1371/journal.pone.0165395
TCGA Expedition: A Data Acquisition and Management System for TCGA Data
Abstract
Background: The Cancer Genome Atlas Project (TCGA) is a National Cancer Institute effort to profile at least 500 cases of 20 different tumor types using genomic platforms and to make these data, both raw and processed, available to all researchers. TCGA data are currently over 1.2 Petabyte in size and include whole genome sequence (WGS), whole exome sequence, methylation, RNA expression, proteomic, and clinical datasets. Publicly accessible TCGA data are released through public portals, but many challenges exist in navigating and using data obtained from these sites. We developed TCGA Expedition to support the research community focused on computational methods for cancer research. Data obtained, versioned, and archived using TCGA Expedition supports command line access at high-performance computing facilities as well as some functionality with third party tools. For a subset of TCGA data collected at University of Pittsburgh, we also re-associate TCGA data with de-identified data from the electronic health records. Here we describe the software as well as the architecture of our repository, methods for loading of TCGA data to multiple platforms, and security and regulatory controls that conform to federal best practices.
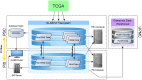
Results: TCGA Expedition software consists of a set of scripts written in Bash, Python and Java that download, extract, harmonize, version and store all TCGA data and metadata. The software generates a versioned, participant- and sample-centered, local TCGA data directory with metadata structures that directly reference the local data files as well as the original data files. The software supports flexible searches of the data via a web portal, user-centric data tracking tools, and data provenance tools. Using this software, we created a collaborative repository, the Pittsburgh Genome Resource Repository (PGRR) that enabled investigators at our institution to work with all TCGA data formats, and to interrogate these data with analysis pipelines, and associated tools. WGS data are especially challenging for individual investigators to use, due to issues with downloading, storage, and processing; having locally accessible WGS BAM files has proven invaluable.
Conclusion: Our open-source, freely available TCGA Expedition software can be used to create a local collaborative infrastructure for acquiring, managing, and analyzing TCGA data and other large public datasets.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Cancer Genome Atlas Project (TCGA) [cited 2015 December 9]. Available from: http://cancergenome.nih.gov/.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous