Combined small angle X-ray solution scattering with atomic force microscopy for characterizing radiation damage on biological macromolecules
- PMID: 27788689
- PMCID: PMC5081678
- DOI: 10.1186/s12900-016-0068-2
Combined small angle X-ray solution scattering with atomic force microscopy for characterizing radiation damage on biological macromolecules
Abstract
Background: Synchrotron radiation facilities are pillars of modern structural biology. Small-Angle X-ray scattering performed at synchrotron sources is often used to characterize the shape of biological macromolecules. A major challenge with high-energy X-ray beam on such macromolecules is the perturbation of sample due to radiation damage.
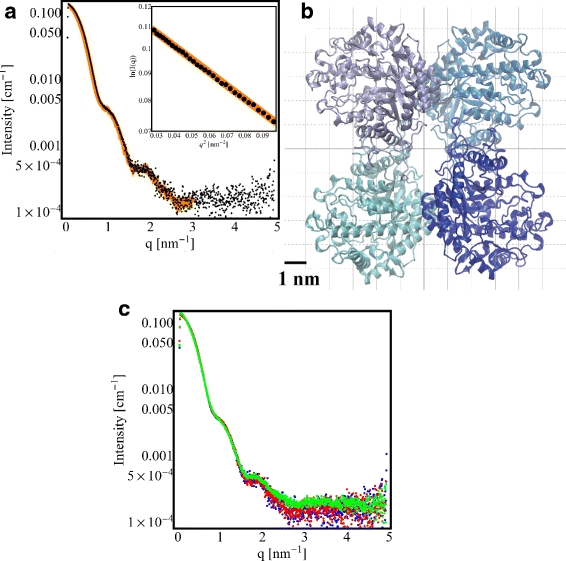
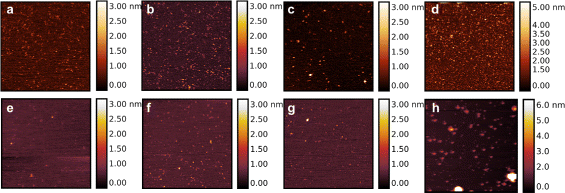
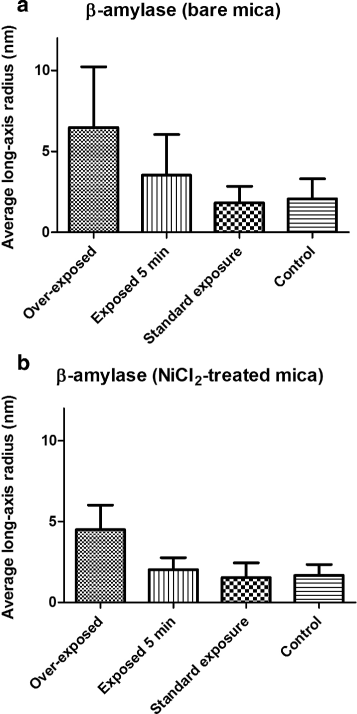
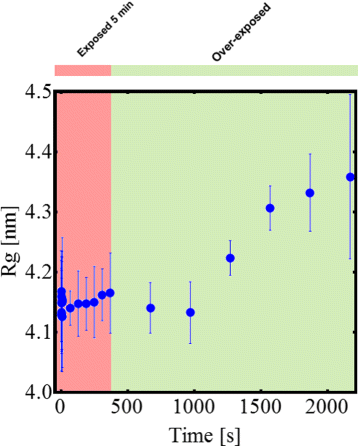
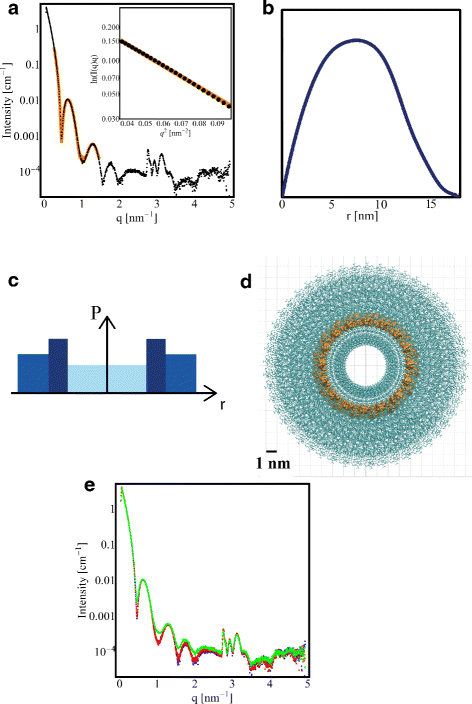
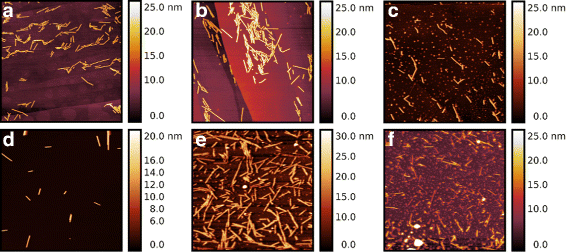
Results: By employing atomic force microscopy, another common technique to determine the shape of biological macromolecules when deposited on flat substrates, we present a protocol to evaluate and characterize consequences of radiation damage. It requires the acquisition of images of irradiated samples at the single molecule level in a timely manner while using minimal amounts of protein. The protocol has been tested on two different molecular systems: a large globular tetremeric enzyme (β-Amylase) and a rod-shape plant virus (tobacco mosaic virus). Radiation damage on the globular enzyme leads to an apparent increase in molecular sizes whereas the effect on the long virus is a breakage into smaller pieces resulting in a decrease of the average long-axis radius.
Conclusions: These results show that radiation damage can appear in different forms and strongly support the need to check the effect of radiation damage at synchrotron sources using the presented protocol.
Keywords: Atomic force microscopy (AFM); Radiation damage; Small angle x-ray scattering (SAXS); Tobacco mosaic virus; β-Amylase.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

