Impact of MYH6 variants in hypoplastic left heart syndrome
- PMID: 27789736
- PMCID: PMC5206387
- DOI: 10.1152/physiolgenomics.00091.2016
Impact of MYH6 variants in hypoplastic left heart syndrome
Abstract
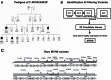
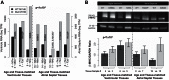
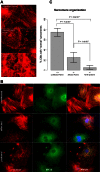
Hypoplastic left heart syndrome (HLHS) is a clinically and anatomically severe form of congenital heart disease (CHD). Although prior studies suggest that HLHS has a complex genetic inheritance, its etiology remains largely unknown. The goal of this study was to characterize a risk gene in HLHS and its effect on HLHS etiology and outcome. We performed next-generation sequencing on a multigenerational family with a high prevalence of CHD/HLHS, identifying a rare variant in the α-myosin heavy chain (MYH6) gene. A case-control study of 190 unrelated HLHS subjects was then performed and compared with the 1000 Genomes Project. Damaging MYH6 variants, including novel, missense, in-frame deletion, premature stop, de novo, and compound heterozygous variants, were significantly enriched in HLHS cases (P < 1 × 10-5). Clinical outcomes analysis showed reduced transplant-free survival in HLHS subjects with damaging MYH6 variants (P < 1 × 10-2). Transcriptome and protein expression analyses with cardiac tissue revealed differential expression of cardiac contractility genes, notably upregulation of the β-myosin heavy chain (MYH7) gene in subjects with MYH6 variants (P < 1 × 10-3). We subsequently used patient-specific induced pluripotent stem cells (iPSCs) to model HLHS in vitro. Early stages of in vitro cardiomyogenesis in iPSCs derived from two unrelated HLHS families mimicked the increased expression of MYH7 observed in vivo (P < 1 × 10-2), while revealing defective cardiomyogenic differentiation. Rare, damaging variants in MYH6 are enriched in HLHS, affect molecular expression of contractility genes, and are predictive of poor outcome. These findings indicate that the etiology of MYH6-associated HLHS can be informed using iPSCs and suggest utility in future clinical applications.
Keywords: cardiomyocyte-autonomous; genetics; hypoplastic left heart syndrome; transplant-free survival outcome; upregulation of contractility genes.
Copyright © 2016 the American Physiological Society.
Figures



References
-
- Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development 130: 6121–6129, 2003. - PubMed
-
- Boneva RS, Botto LD, Moore CA, Yang Q, Correa A, Erickson JD. Mortality associated with congenital heart defects in the United States: trends and racial disparities, 1979–1997. Circulation 103: 2376–2381, 2001. - PubMed
-
- Carniel E, Taylor MR, Sinagra G, Di Lenarda A, Ku L, Fain PR, Boucek MM, Cavanaugh J, Miocic S, Slavov D, Graw SL, Feiger J, Zhu XZ, Dao D, Ferguson DA, Bristow MR, Mestroni L. Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation 112: 54–59, 2005. - PubMed
-
- Chen J, Kubalak SW, Minamisawa S, Price RL, Becker KD, Hickey R, Ross J Jr, Chien KR. Selective requirement of myosin light chain 2v in embryonic heart function. J Biol Chem 273: 1252–1256, 1998. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

