Plant-Specific Preprotein and Amino Acid Transporter Proteins Are Required for tRNA Import into Mitochondria
- PMID: 27789739
- PMCID: PMC5129730
- DOI: 10.1104/pp.16.01519
Plant-Specific Preprotein and Amino Acid Transporter Proteins Are Required for tRNA Import into Mitochondria
Abstract
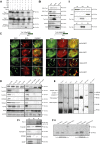
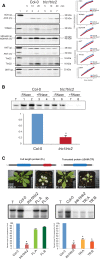
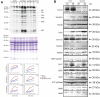
A variety of eukaryotes, in particular plants, do not contain the required number of tRNAs to support the translation of mitochondria-encoded genes and thus need to import tRNAs from the cytosol. This study identified two Arabidopsis (Arabidopsis thaliana) proteins, Tric1 and Tric2 (for tRNA import component), which on simultaneous inactivation by T-DNA insertion lines displayed a severely delayed and chlorotic growth phenotype and significantly reduced tRNA import capacity into isolated mitochondria. The predicted tRNA-binding domain of Tric1 and Tric2, a sterile-α-motif at the C-terminal end of the protein, was required to restore tRNA uptake ability in mitochondria of complemented plants. The purified predicted tRNA-binding domain binds the T-arm of the tRNA for alanine with conserved lysine residues required for binding. T-DNA inactivation of both Tric proteins further resulted in an increase in the in vitro rate of in organello protein synthesis, which was mediated by a reorganization of the nuclear transcriptome, in particular of genes encoding a variety of proteins required for mitochondrial gene expression at both the transcriptional and translational levels. The characterization of Tric1/2 provides mechanistic insight into the process of tRNA import into mitochondria and supports the theory that the tRNA import pathway resulted from the repurposing of a preexisting protein import apparatus.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures





Similar articles
-
Structural analysis of the SAM domain of the Arabidopsis mitochondrial tRNA import receptor.J Biol Chem. 2024 May;300(5):107258. doi: 10.1016/j.jbc.2024.107258. Epub 2024 Apr 4. J Biol Chem. 2024. PMID: 38582448 Free PMC article.
-
In vitro RNA uptake studies in plant mitochondria.Methods Mol Biol. 2015;1305:45-60. doi: 10.1007/978-1-4939-2639-8_4. Methods Mol Biol. 2015. PMID: 25910726
-
Functional definition of outer membrane proteins involved in preprotein import into mitochondria.Plant Cell. 2007 Nov;19(11):3739-59. doi: 10.1105/tpc.107.050534. Epub 2007 Nov 2. Plant Cell. 2007. PMID: 17981999 Free PMC article.
-
Mitochondrial tRNA import and its consequences for mitochondrial translation.Annu Rev Biochem. 2011;80:1033-53. doi: 10.1146/annurev-biochem-060109-092838. Annu Rev Biochem. 2011. PMID: 21417719 Review.
-
Recent advances in tRNA mitochondrial import.Trends Biochem Sci. 2008 Jul;33(7):320-9. doi: 10.1016/j.tibs.2008.04.010. Epub 2008 May 29. Trends Biochem Sci. 2008. PMID: 18513973 Review.
Cited by
-
Creativity comes from interactions: modules of protein interactions in plants.FEBS J. 2022 Mar;289(6):1492-1514. doi: 10.1111/febs.15847. Epub 2021 May 1. FEBS J. 2022. PMID: 33774929 Free PMC article. Review.
-
Tric proteins and TOM complex subunits are involved in the import of short DNA fragments into Arabidopsis mitochondria.Protoplasma. 2025 Jun 24. doi: 10.1007/s00709-025-02083-9. Online ahead of print. Protoplasma. 2025. PMID: 40555883
-
Defying gravity: WEEP promotes negative gravitropism in peach trees by establishing asymmetric auxin gradients.Plant Physiol. 2024 May 31;195(2):1229-1255. doi: 10.1093/plphys/kiae085. Plant Physiol. 2024. PMID: 38366651 Free PMC article.
-
The plastome of Melocactus glaucescens Buining & Brederoo reveals unique evolutionary features and loss of essential tRNA genes.Planta. 2022 Feb 3;255(3):57. doi: 10.1007/s00425-022-03841-2. Planta. 2022. PMID: 35113261
-
Photosynthetic demands on translational machinery drive retention of redundant tRNA metabolism in plant organelles.Proc Natl Acad Sci U S A. 2024 Dec 24;121(52):e2421485121. doi: 10.1073/pnas.2421485121. Epub 2024 Dec 18. Proc Natl Acad Sci U S A. 2024. PMID: 39693336 Free PMC article.
References
-
- Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395 - PubMed
-
- Altschul SF, Koonin EV (1998) Iterated profile searches with PSI-BLAST: a tool for discovery in protein databases. Trends Biochem Sci 23: 444–447 - PubMed
-
- Aronsson H, Jarvis RP (2011) Rapid isolation of Arabidopsis chloroplasts and their use for in vitro protein import assays. Methods Mol Biol 774: 281–305 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

