The bacterial enhancer-dependent RNA polymerase
- PMID: 27789741
- PMCID: PMC5095919
- DOI: 10.1042/BCJ20160741C
The bacterial enhancer-dependent RNA polymerase
Abstract
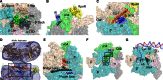
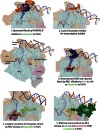
Transcription initiation is highly regulated in bacterial cells, allowing adaptive gene regulation in response to environment cues. One class of promoter specificity factor called sigma54 enables such adaptive gene expression through its ability to lock the RNA polymerase down into a state unable to melt out promoter DNA for transcription initiation. Promoter DNA opening then occurs through the action of specialized transcription control proteins called bacterial enhancer-binding proteins (bEBPs) that remodel the sigma54 factor within the closed promoter complexes. The remodelling of sigma54 occurs through an ATP-binding and hydrolysis reaction carried out by the bEBPs. The regulation of bEBP self-assembly into typically homomeric hexamers allows regulated gene expression since the self-assembly is required for bEBP ATPase activity and its direct engagement with the sigma54 factor during the remodelling reaction. Crystallographic studies have now established that in the closed promoter complex, the sigma54 factor occupies the bacterial RNA polymerase in ways that will physically impede promoter DNA opening and the loading of melted out promoter DNA into the DNA-binding clefts of the RNA polymerase. Large-scale structural re-organizations of sigma54 require contact of the bEBP with an amino-terminal glutamine and leucine-rich sequence of sigma54, and lead to domain movements within the core RNA polymerase necessary for making open promoter complexes and synthesizing the nascent RNA transcript.
Keywords: AAA+ proteins; RNA polymerase; bEBPs; sigma54; sigma70; transcription.
© 2016 The Author(s).
Figures


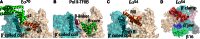

Similar articles
-
Subunit specialization in AAA+ proteins and substrate unfolding during transcription complex remodeling.Proc Natl Acad Sci U S A. 2025 Apr 29;122(17):e2425868122. doi: 10.1073/pnas.2425868122. Epub 2025 Apr 24. Proc Natl Acad Sci U S A. 2025. PMID: 40273105 Free PMC article.
-
Bacterial enhancer-binding proteins: unlocking sigma54-dependent gene transcription.Curr Opin Struct Biol. 2007 Feb;17(1):110-6. doi: 10.1016/j.sbi.2006.11.002. Epub 2006 Dec 6. Curr Opin Struct Biol. 2007. PMID: 17157497 Review.
-
Correlating protein footprinting with mutational analysis in the bacterial transcription factor sigma54 (sigmaN).Nucleic Acids Res. 2002 Feb 15;30(4):1016-28. doi: 10.1093/nar/30.4.1016. Nucleic Acids Res. 2002. PMID: 11842114 Free PMC article.
-
The sigma 54 DNA-binding domain includes a determinant of enhancer responsiveness.Mol Microbiol. 1999 Sep;33(6):1200-9. doi: 10.1046/j.1365-2958.1999.01566.x. Mol Microbiol. 1999. PMID: 10510234
-
Bacterial Enhancer Binding Proteins-AAA+ Proteins in Transcription Activation.Biomolecules. 2020 Feb 25;10(3):351. doi: 10.3390/biom10030351. Biomolecules. 2020. PMID: 32106553 Free PMC article. Review.
Cited by
-
The sigma factor σ54 is required for the long-term survival of Leptospira biflexa in water.Mol Microbiol. 2018 Apr 6:10.1111/mmi.13967. doi: 10.1111/mmi.13967. Online ahead of print. Mol Microbiol. 2018. PMID: 29633391 Free PMC article.
-
Analysis and comparison of the bacterial σ54 regulon: Evidence of phylogenetic trends in gene regulation.PLoS One. 2025 Aug 1;20(8):e0327805. doi: 10.1371/journal.pone.0327805. eCollection 2025. PLoS One. 2025. PMID: 40748940 Free PMC article.
-
Chromosome organization in bacteria: mechanistic insights into genome structure and function.Nat Rev Genet. 2020 Apr;21(4):227-242. doi: 10.1038/s41576-019-0185-4. Epub 2019 Nov 25. Nat Rev Genet. 2020. PMID: 31767998 Review.
-
Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology.FEMS Microbiol Rev. 2019 May 1;43(3):304-339. doi: 10.1093/femsre/fuz001. FEMS Microbiol Rev. 2019. PMID: 30721976 Free PMC article. Review.
-
Subunit specialization in AAA+ proteins and substrate unfolding during transcription complex remodeling.Proc Natl Acad Sci U S A. 2025 Apr 29;122(17):e2425868122. doi: 10.1073/pnas.2425868122. Epub 2025 Apr 24. Proc Natl Acad Sci U S A. 2025. PMID: 40273105 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

