Systems-level analysis of mechanisms regulating yeast metabolic flux
- PMID: 27789812
- PMCID: PMC5414049
- DOI: 10.1126/science.aaf2786
Systems-level analysis of mechanisms regulating yeast metabolic flux
Abstract
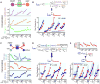
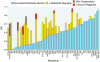
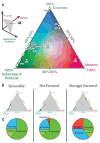
Cellular metabolic fluxes are determined by enzyme activities and metabolite abundances. Biochemical approaches reveal the impact of specific substrates or regulators on enzyme kinetics but do not capture the extent to which metabolite and enzyme concentrations vary across physiological states and, therefore, how cellular reactions are regulated. We measured enzyme and metabolite concentrations and metabolic fluxes across 25 steady-state yeast cultures. We then assessed the extent to which flux can be explained by a Michaelis-Menten relationship between enzyme, substrate, product, and potential regulator concentrations. This revealed three previously unrecognized instances of cross-pathway regulation, which we biochemically verified. One of these involved inhibition of pyruvate kinase by citrate, which accumulated and thereby curtailed glycolytic outflow in nitrogen-limited yeast. Overall, substrate concentrations were the strongest driver of the net rates of cellular metabolic reactions, with metabolite concentrations collectively having more than double the physiological impact of enzymes.
Copyright © 2016, American Association for the Advancement of Science.
Figures



Comment in
-
Metabolism-Centric Trans-Omics.Cell Syst. 2017 Jan 25;4(1):19-20. doi: 10.1016/j.cels.2017.01.007. Cell Syst. 2017. PMID: 28125790
References
-
- Kacser H, Burns JA. Symposia of the Society for Experimental Biology. 1973;27:65. - PubMed
-
- Fell D. Understanding the control of metabolism. Portland Press Ltd; 1997.
-
- Tummler K, Lubitz T, Schelker M, Klipp E. The FEBS journal. 2014;281:549. - PubMed
-
- Hauf J, Zimmermann FK, Muller S. Enzyme and microbial technology. 2000;26:688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

