Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER
- PMID: 27789813
- PMCID: PMC6528812
- DOI: 10.1126/science.aaf3928
Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER
Abstract
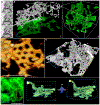
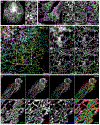
The endoplasmic reticulum (ER) is an expansive, membrane-enclosed organelle that plays crucial roles in numerous cellular functions. We used emerging superresolution imaging technologies to clarify the morphology and dynamics of the peripheral ER, which contacts and modulates most other intracellular organelles. Peripheral components of the ER have classically been described as comprising both tubules and flat sheets. We show that this system consists almost exclusively of tubules at varying densities, including structures that we term ER matrices. Conventional optical imaging technologies had led to misidentification of these structures as sheets because of the dense clustering of tubular junctions and a previously uncharacterized rapid form of ER motion. The existence of ER matrices explains previous confounding evidence that had indicated the occurrence of ER "sheet" proliferation after overexpression of tubular junction-forming proteins.
Copyright © 2016, American Association for the Advancement of Science.
Figures





Comment in
-
A finer look at a fine cellular meshwork.Science. 2016 Oct 28;354(6311):415-416. doi: 10.1126/science.aal0052. Science. 2016. PMID: 27789827 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

