Transcriptome Analysis of a New Peanut Seed Coat Mutant for the Physiological Regulatory Mechanism Involved in Seed Coat Cracking and Pigmentation
- PMID: 27790222
- PMCID: PMC5063860
- DOI: 10.3389/fpls.2016.01491
Transcriptome Analysis of a New Peanut Seed Coat Mutant for the Physiological Regulatory Mechanism Involved in Seed Coat Cracking and Pigmentation
Abstract
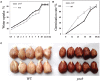
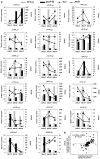
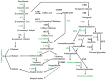
Seed-coat cracking and undesirable color of seed coat highly affects external appearance and commercial value of peanuts (Arachis hypogaea L.). With an objective to find genetic solution to the above problems, a peanut mutant with cracking and brown colored seed coat (testa) was identified from an EMS treated mutant population and designated as "peanut seed coat crack and brown color mutant line (pscb)." The seed coat weight of the mutant was almost twice of the wild type, and the germination time was significantly shorter than wild type. Further, the mutant had lower level of lignin, anthocyanin, proanthocyanidin content, and highly increased level of melanin content as compared to wild type. Using RNA-Seq, we examined the seed coat transcriptome in three stages of seed development in the wild type and the pscb mutant. The RNA-Seq analysis revealed presence of highly differentially expressed phenylpropanoid and flavonoid pathway genes in all the three seed development stages, especially at 40 days after flowering (DAF40). Also, the expression of polyphenol oxidases and peroxidase were found to be activated significantly especially in the late seed developmental stage. The genome-wide comparative study of the expression profiles revealed 62 differentially expressed genes common across all the three stages. By analyzing the expression patterns and the sequences of the common differentially expressed genes of the three stages, three candidate genes namely c36498_g1 (CCoAOMT1), c40902_g2 (kinesin), and c33560_g1 (MYB3) were identified responsible for seed-coat cracking and brown color phenotype. Therefore, this study not only provided candidate genes but also provided greater insights and molecular genetic control of peanut seed-coat cracking and color variation. The information generated in this study will facilitate further identification of causal gene and diagnostic markers for breeding improved peanut varieties with smooth and desirable seed coat color.
Keywords: RNA-seq; flavonoid pathway; peanut (Arachis hypogaea); pigmentation; seed-coat cracking.
Figures





References
-
- Agarwal K., Menon K. (1974). Lignin content and seed coat thickness in relation to seed coat cracking in soybean. Seed Res. 2, 64–66.
-
- Appelhagen I., Lu G. H., Huep G., Schmelzer E., Weisshaar B., Sagasser M. (2011). TRANSPARENT TESTA1 interacts with R2R3-MYB factors and affects early and late steps of flavonoid biosynthesis in the endothelium of Arabidopsis thaliana seeds. Plant J. 67, 406–419. 10.1111/j.1365-313X.2011.04603.x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

