One step forwards for the routine use of high-throughput DNA sequencing in environmental monitoring. An efficient and standardizable method to maximize the detection of environmental bacteria
- PMID: 27790854
- PMCID: PMC5300880
- DOI: 10.1002/mbo3.421
One step forwards for the routine use of high-throughput DNA sequencing in environmental monitoring. An efficient and standardizable method to maximize the detection of environmental bacteria
Abstract
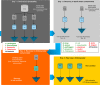
We propose an innovative, repeatable, and reliable experimental workflow to concentrate and detect environmental bacteria in drinking water using molecular techniques. We first concentrated bacteria in water samples using tangential flow filtration and then we evaluated two methods of environmental DNA extraction. We performed tests on both artificially contaminated water samples and real drinking water samples. The efficiency of the experimental workflow was measured through qPCR. The successful applicability of the high-throughput DNA sequencing (HTS) approach was demonstrated on drinking water samples. Our results demonstrate the feasibility of our approach in high-throughput-based studies, and we suggest incorporating it in monitoring strategies to have a better representation of the microbial community. In the recent years, HTS techniques have become key tools in the study of microbial communities. To make the leap from academic laboratories to the routine monitoring (e.g., water treatment plants laboratories), we here propose an experimental workflow suitable for the introduction of HTS as a standard method for detecting environmental bacteria.
Keywords: qPCR; DNA extraction; complex matrix; environmental bacteria; high-throughput sequencing; next-generation sequencing; tangential flow concentration; water.
© 2016 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures

References
-
- Alexander, J. , Bollmann, A. , Seitz, W. , & Schwartz, T. (2015). Microbiological characterization of aquatic microbiomes targeting taxonomical marker genes and antibiotic resistance genes of opportunistic bacteria. Science of the Total Environment, 512, 316–325. - PubMed
-
- Ashbolt, N. J. , Grabow, W. O. K. , & Snozzi, M. (2001). Indicators of microbial water quality. In: Fewtrell L., Bartram J. (Eds.), Water Quality: Guidelines, Standards and Health. Risk assessment and management for water‐related infectious disease. London: IWA Publishing; (Chapter 13), pp. 289–315.
-
- Berendonk, T. U. , Manaia, C. M. , Merlin, C. , Fatta‐Kassinos, D. , Cytryn, E. , Walsh, F. , … Martinez, J. L. (2015). Tackling antibiotic resistance: The environmental framework. Nature Reviews Microbiology, 13, 310–317. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

