Extracellular matrix stiffness causes systematic variations in proliferation and chemosensitivity in myeloid leukemias
- PMID: 27790998
- PMCID: PMC5086998
- DOI: 10.1073/pnas.1611338113
Extracellular matrix stiffness causes systematic variations in proliferation and chemosensitivity in myeloid leukemias
Abstract
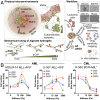
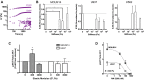
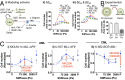
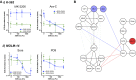
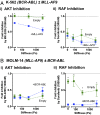
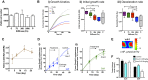
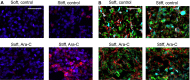
Extracellular matrix stiffness influences biological functions of some tumors. However, it remains unclear how cancer subtypes with different oncogenic mutations respond to matrix stiffness. In addition, the relevance of matrix stiffness to in vivo tumor growth kinetics and drug efficacy remains elusive. Here, we designed 3D hydrogels with physical parameters relevant to hematopoietic tissues and adapted them to a quantitative high-throughput screening format to facilitate mechanistic investigations into the role of matrix stiffness on myeloid leukemias. Matrix stiffness regulates proliferation of some acute myeloid leukemia types, including MLL-AF9+ MOLM-14 cells, in a biphasic manner by autocrine regulation, whereas it decreases that of chronic myeloid leukemia BCR-ABL+ K-562 cells. Although Arg-Gly-Asp (RGD) integrin ligand and matrix softening confer resistance to a number of drugs, cells become sensitive to drugs against protein kinase B (PKB or AKT) and rapidly accelerated fibrosarcoma (RAF) proteins regardless of matrix stiffness when MLL-AF9 and BCR-ABL are overexpressed in K-562 and MOLM-14 cells, respectively. By adapting the same hydrogels to a xenograft model of extramedullary leukemias, we confirm the pathological relevance of matrix stiffness in growth kinetics and drug sensitivity against standard chemotherapy in vivo. The results thus demonstrate the importance of incorporating 3D mechanical cues into screening for anticancer drugs.
Keywords: biomaterials; cancer; drug screening; matrix stiffness; systems pharmacology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Daley GQ. Chronic myeloid leukemia: Proving ground for cancer stem cells. Cell. 2004;119(3):314–316. - PubMed
-
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7(11):823–833. - PubMed
-
- Cheng JQ, Lindsley CW, Cheng GZ, Yang H, Nicosia SV. The Akt/PKB pathway: Molecular target for cancer drug discovery. Oncogene. 2005;24(50):7482–7492. - PubMed
-
- Chaudhuri O, et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater. 2014;13(10):970–978. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

