Differential roles of the RNases H in preventing chromosome instability
- PMID: 27791008
- PMCID: PMC5086985
- DOI: 10.1073/pnas.1613448113
Differential roles of the RNases H in preventing chromosome instability
Abstract
DNA:RNA hybrids can lead to DNA damage and genome instability. This damage can be prevented by degradation of the RNA in the hybrid by two evolutionarily conserved enzymes, RNase H1 and H2. Indeed, RNase H-deficient cells have increased chromosomal rearrangements. However, the quantitative and spatial contributions of the individual enzymes to hybrid removal have been unclear. Additionally, RNase H2 can remove single ribonucleotides misincorporated into DNA during replication. The relative contribution of DNA:RNA hybrids and misincorporated ribonucleotides to chromosome instability also was uncertain. To address these issues, we studied the frequency and location of loss-of-heterozygosity (LOH) events on chromosome III in Saccharomyces cerevisiae strains that were defective for RNase H1, H2, or both. We showed that RNase H2 plays the major role in preventing chromosome III instability through its hybrid-removal activity. Furthermore, RNase H2 acts pervasively at many hybrids along the chromosome. In contrast, RNase H1 acts to prevent LOH within a small region of chromosome III where the instability is dependent upon two hybrid-prone sequences. This restriction of RNase H1 activity to a subset of hybrids is not the result of its constrained localization, because we found it at hybrids genome-wide. This result suggests that the genome-protection activity of RNase H1 is regulated at a step after hybrid recognition. The global function of RNase H2 and the region-specific function of RNase H1 provide insight into why these enzymes with overlapping hybrid-removal activities have been conserved throughout evolution.
Keywords: DNA:RNA hybrids; R-loops; RNase H; chromosome instability; genome instability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






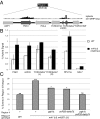


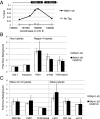

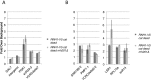
References
-
- Santos-Pereira JM, Aguilera A. R loops: New modulators of genome dynamics and function. Nat Rev Genet. 2015;16(10):583–597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

