A conserved αβ transmembrane interface forms the core of a compact T-cell receptor-CD3 structure within the membrane
- PMID: 27791034
- PMCID: PMC5086997
- DOI: 10.1073/pnas.1611445113
A conserved αβ transmembrane interface forms the core of a compact T-cell receptor-CD3 structure within the membrane
Abstract
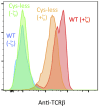
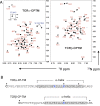
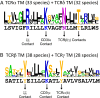
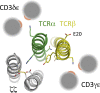
The T-cell antigen receptor (TCR) is an assembly of eight type I single-pass membrane proteins that occupies a central position in adaptive immunity. Many TCR-triggering models invoke an alteration in receptor complex structure as the initiating event, but both the precise subunit organization and the pathway by which ligand-induced alterations are transferred to the cytoplasmic signaling domains are unknown. Here, we show that the receptor complex transmembrane (TM) domains form an intimately associated eight-helix bundle organized by a specific interhelical TCR TM interface. The salient features of this core structure are absolutely conserved between αβ and γδ TCR sequences and throughout vertebrate evolution, and mutations at key interface residues caused defects in the formation of stable TCRαβ:CD3δε:CD3γε:ζζ complexes. These findings demonstrate that the eight TCR-CD3 subunits form a compact and precisely organized structure within the membrane and provide a structural basis for further investigation of conformationally regulated models of transbilayer TCR signaling.
Keywords: MD simulation; NMR; T-cell receptor; cysteine cross-linking; transmembrane structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures















References
-
- Beddoe T, et al. Antigen ligation triggers a conformational change within the constant domain of the αβ T cell receptor. Immunity. 2009;30(6):777–788. - PubMed
-
- Kuhns MS, Davis MM. Disruption of extracellular interactions impairs T cell receptor-CD3 complex stability and signaling. Immunity. 2007;26(3):357–369. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

