miRNA92a targets KLF2 and the phosphatase PTEN signaling to promote human T follicular helper precursors in T1D islet autoimmunity
- PMID: 27791035
- PMCID: PMC5087025
- DOI: 10.1073/pnas.1606646113
miRNA92a targets KLF2 and the phosphatase PTEN signaling to promote human T follicular helper precursors in T1D islet autoimmunity
Erratum in
-
Correction for Serr et al., miRNA92a targets KLF2 and the phosphatase PTEN signaling to promote human T follicular helper precursors in T1D islet autoimmunity.Proc Natl Acad Sci U S A. 2018 Apr 24;115(17):E4142. doi: 10.1073/pnas.1805675115. Epub 2018 Apr 16. Proc Natl Acad Sci U S A. 2018. PMID: 29666264 Free PMC article. No abstract available.
Abstract
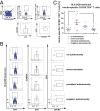
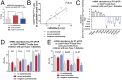
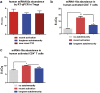
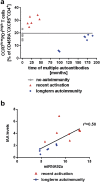
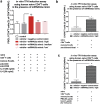
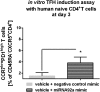
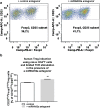
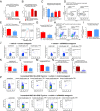
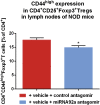
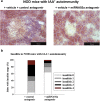
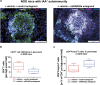
Aberrant immune activation mediated by T effector cell populations is pivotal in the onset of autoimmunity in type 1 diabetes (T1D). T follicular helper (TFH) cells are essential in the induction of high-affinity antibodies, and their precursor memory compartment circulates in the blood. The role of TFH precursors in the onset of islet autoimmunity and signaling pathways regulating their differentiation is incompletely understood. Here, we provide direct evidence that during onset of islet autoimmunity, the insulin-specific target T-cell population is enriched with a C-X-C chemokine receptor type 5 (CXCR5)+CD4+ TFH precursor phenotype. During onset of islet autoimmunity, the frequency of TFH precursors was controlled by high expression of microRNA92a (miRNA92a). miRNA92a-mediated TFH precursor induction was regulated by phosphatase and tension homolog (PTEN) - phosphoinositol-3-kinase (PI3K) signaling involving PTEN and forkhead box protein O1 (Foxo1), supporting autoantibody generation and triggering the onset of islet autoimmunity. Moreover, we identify Krueppel-like factor 2 (KLF2) as a target of miRNA92a in regulating human TFH precursor induction. Importantly, a miRNA92a antagomir completely blocked induction of human TFH precursors in vitro. More importantly, in vivo application of a miRNA92a antagomir to nonobese diabetic (NOD) mice with ongoing islet autoimmunity resulted in a significant reduction of TFH precursors in peripheral blood and pancreatic lymph nodes. Moreover, miRNA92a antagomir application reduced immune infiltration and activation in pancreata of NOD mice as well as humanized NOD Scid IL2 receptor gamma chain knockout (NSG) human leucocyte antigen (HLA)-DQ8 transgenic animals. We therefore propose that miRNA92a and the PTEN-PI3K-KLF2 signaling network could function as targets for innovative precision medicines to reduce T1D islet autoimmunity.
Keywords: KLF2; PTEN-PI3K signaling; T follicular helper cells; miRNA92a; type 1 diabetes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Unanue ER. Antigen presentation in the autoimmune diabetes of the NOD mouse. Annu Rev Immunol. 2014;32:579–608. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

