Functions and regulation of the Brr2 RNA helicase during splicing
- PMID: 27792457
- PMCID: PMC5224448
- DOI: 10.1080/15384101.2016.1249549
Functions and regulation of the Brr2 RNA helicase during splicing
Abstract
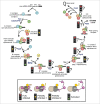
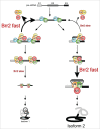
Pre-mRNA splicing entails the stepwise assembly of an inactive spliceosome, its catalytic activation, splicing catalysis and spliceosome disassembly. Transitions in this reaction cycle are accompanied by compositional and conformational rearrangements of the underlying RNA-protein interaction networks, which are driven and controlled by 8 conserved superfamily 2 RNA helicases. The Ski2-like helicase, Brr2, provides the key remodeling activity during spliceosome activation and is additionally implicated in the catalytic and disassembly phases of splicing, indicating that Brr2 needs to be tightly regulated during splicing. Recent structural and functional analyses have begun to unravel how Brr2 regulation is established via multiple layers of intra- and inter-molecular mechanisms. Brr2 has an unusual structure, including a long N-terminal region and a catalytically inactive C-terminal helicase cassette, which can auto-inhibit and auto-activate the enzyme, respectively. Both elements are essential, also serve as protein-protein interaction devices and the N-terminal region is required for stable Brr2 association with the tri-snRNP, tri-snRNP stability and retention of U5 and U6 snRNAs during spliceosome activation in vivo. Furthermore, a C-terminal region of the Prp8 protein, comprising consecutive RNase H-like and Jab1/MPN-like domains, can both up- and down-regulate Brr2 activity. Biochemical studies revealed an intricate cross-talk among the various cis- and trans-regulatory mechanisms. Comparison of isolated Brr2 to electron cryo-microscopic structures of yeast and human U4/U6•U5 tri-snRNPs and spliceosomes indicates how some of the regulatory elements exert their functions during splicing. The various modulatory mechanisms acting on Brr2 might be exploited to enhance splicing fidelity and to regulate alternative splicing.
Keywords: Pre-mRNA splicing; RNA helicase structure,; function and regulation; regulation of pre-mRNA splicing; remodeling of RNA-protein complexes; spliceosome catalytic activation.
Figures

Similar articles
-
The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation.Genes Dev. 2015 Dec 15;29(24):2576-87. doi: 10.1101/gad.271528.115. Epub 2015 Dec 4. Genes Dev. 2015. PMID: 26637280 Free PMC article.
-
Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2.Cell Cycle. 2017 Jan 2;16(1):100-112. doi: 10.1080/15384101.2016.1255384. Epub 2016 Nov 23. Cell Cycle. 2017. PMID: 27880071 Free PMC article.
-
The inactive C-terminal cassette of the dual-cassette RNA helicase BRR2 both stimulates and inhibits the activity of the N-terminal helicase unit.J Biol Chem. 2020 Feb 14;295(7):2097-2112. doi: 10.1074/jbc.RA119.010964. Epub 2019 Dec 30. J Biol Chem. 2020. PMID: 31914407 Free PMC article.
-
Novel regulatory principles of the spliceosomal Brr2 RNA helicase and links to retinal disease in humans.RNA Biol. 2014;11(4):298-312. doi: 10.4161/rna.28353. Epub 2014 Mar 5. RNA Biol. 2014. PMID: 24643059 Free PMC article. Review.
-
Multiple genetic and biochemical interactions of Brr2, Prp8, Prp31, Prp1 and Prp4 kinase suggest a function in the control of the activation of spliceosomes in Schizosaccharomyces pombe.Curr Genet. 2005 Sep;48(3):151-61. doi: 10.1007/s00294-005-0013-6. Epub 2005 Oct 12. Curr Genet. 2005. PMID: 16133344 Review.
Cited by
-
The interaction of DNA repair factors ASCC2 and ASCC3 is affected by somatic cancer mutations.Nat Commun. 2020 Nov 2;11(1):5535. doi: 10.1038/s41467-020-19221-x. Nat Commun. 2020. PMID: 33139697 Free PMC article.
-
An Allosteric Network for Spliceosome Activation Revealed by High-Throughput Suppressor Analysis in Saccharomyces cerevisiae.Genetics. 2019 May;212(1):111-124. doi: 10.1534/genetics.119.301922. Epub 2019 Mar 21. Genetics. 2019. PMID: 30898770 Free PMC article.
-
RNA and Proteins: Mutual Respect.F1000Res. 2017 Mar 27;6:345. doi: 10.12688/f1000research.10572.1. eCollection 2017. F1000Res. 2017. PMID: 28408981 Free PMC article. Review.
-
Ribosomal protein S7 ubiquitination during ER stress in yeast is associated with selective mRNA translation and stress outcome.Sci Rep. 2020 Nov 12;10(1):19669. doi: 10.1038/s41598-020-76239-3. Sci Rep. 2020. PMID: 33184379 Free PMC article.
-
Large-scale map of RNA-binding protein interactomes across the mRNA life cycle.Mol Cell. 2024 Oct 3;84(19):3790-3809.e8. doi: 10.1016/j.molcel.2024.08.030. Epub 2024 Sep 19. Mol Cell. 2024. PMID: 39303721
References
-
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNAs by the spliceosome In: Gesteland RF, Atkins JF, eds. The RNA world First edition. Cold Spring Harbor, NY: Cold Spring Harbor Labratory Press, 1993:303-57
-
- Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011; 3:1-24; http://dx.doi.org/10.1101/cshperspect.a003707 - DOI - PMC - PubMed
-
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009; 136:701-18; PMID:19239890; http://dx.doi.org/10.1016/j.cell.2009.02.009 - DOI - PubMed
-
- Will CL, Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 2001; 13:290-301; PMID:11343899; http://dx.doi.org/10.1016/S0955-0674(00)00211-8 - DOI - PubMed
-
- Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet 2002; 36:333-60; PMID:12429696; http://dx.doi.org/10.1146/annurev.genet.36.043002.091635 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
