Hedgehog pathway dysregulation contributes to the pathogenesis of human gastrointestinal stromal tumors via GLI-mediated activation of KIT expression
- PMID: 27793025
- PMCID: PMC5346634
- DOI: 10.18632/oncotarget.12909
Hedgehog pathway dysregulation contributes to the pathogenesis of human gastrointestinal stromal tumors via GLI-mediated activation of KIT expression
Abstract
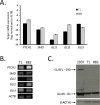
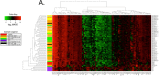
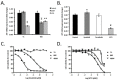
Gastrointestinal stromal tumors (GIST) arise within the interstitial cell of Cajal (ICC) lineage due to activating KIT/PDGFRA mutations. Both ICC and GIST possess primary cilia (PC), which coordinate PDGFRA and Hedgehog signaling, regulators of gastrointestinal mesenchymal development. Therefore, we hypothesized that Hedgehog signaling may be altered in human GIST and controls KIT expression. Quantitative RT-PCR, microarrays, and next generation sequencing were used to describe Hedgehog/PC-related genes in purified human ICC and GIST. Genetic and pharmacologic approaches were employed to investigate the effects of GLI manipulation on KIT expression and GIST cell viability. We report that Hedgehog pathway and PC components are expressed in ICC and GIST and subject to dysregulation during GIST oncogenesis, irrespective of KIT/PDGFRA mutation status. Using genomic profiling, 10.2% of 186 GIST studied had potentially deleterious genomic alterations in 5 Hedgehog-related genes analyzed, including in the PTCH1 tumor suppressor (1.6%). Expression of the predominantly repressive GLI isoform, GLI3, was inversely correlated with KIT mRNA levels in GIST cells and non-KIT/non-PDGFRA mutant GIST. Overexpression of the 83-kDa repressive form of GLI3 or small interfering RNA-mediated knockdown of the activating isoforms GLI1/2 reduced KIT mRNA. Treatment with GLI1/2 inhibitors, including arsenic trioxide, significantly increased GLI3 binding to the KIT promoter, decreased KIT expression, and reduced viability in imatinib-sensitive and imatinib-resistant GIST cells. These data offer new evidence that genes necessary for Hedgehog signaling and PC function in ICC are dysregulated in GIST. Hedgehog signaling activates KIT expression irrespective of mutation status, offering a novel approach to treat imatinib-resistant GIST.
Keywords: GIST; GLI; ICC; arsenic trioxide; imatinib-resistant.
Conflict of interest statement
Juliann Chmielecki, Kai Wang, Deborah Morosini, and Jeffrey Ross are employees of and equity holders in Foundation Medicine, Inc., the provider of the FoundationOne™ and FoundationOne Heme™ assays utilized in this study. Jason Sicklick receives research funds from Foundation Medicine, Inc., Novartis Pharmaceuticals, and Blueprint Medicines. The other authors have nothing to disclose.
Figures
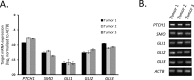




References
-
- Bardsley MR, Horvath VJ, Asuzu DT, Lorincz A, Redelman D, Hayashi Y, Popko LN, Young DL, Lomberk GA, Urrutia RA, Farrugia G, Rubin BP, Ordog T. Kitlow stem cells cause resistance to Kit/platelet-derived growth factor alpha inhibitors in murine gastrointestinal stromal tumors. Gastroenterology. 2010;139:942–52. doi: 10.1053/j.gastro.2010.05.083. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

