Left-right axis asymmetry determining human Cryptic gene is transcriptionally repressed by Snail
- PMID: 27793090
- PMCID: PMC5084438
- DOI: 10.1186/s12861-016-0141-x
Left-right axis asymmetry determining human Cryptic gene is transcriptionally repressed by Snail
Abstract
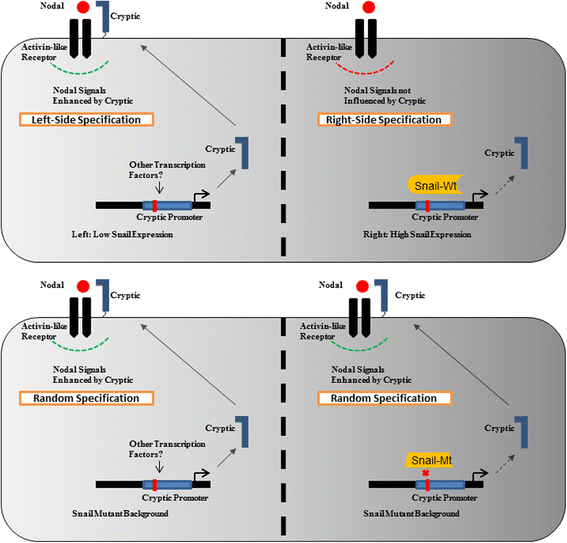
Background: Establishment of the left-right axis is important for positioning organs asymmetrically in the developing vertebrate-embryo. A number of factors like maternally deposited molecules have emerged essential in initiating the specification of the axis; the downstream events, however, are regulated by signal-transduction and gene-expression changes identifying which remains a crucial challenge. The EGF-CFC family member Cryptic, that functions as a co-receptor for some TGF-beta ligands, is developmentally expressed in higher mammals and mutations in the gene cause loss or change in left-right axis asymmetry. Despite the strong phenotype, no transcriptional-regulator of this gene is known till date.
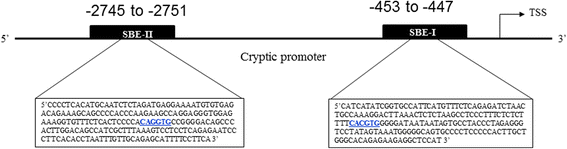
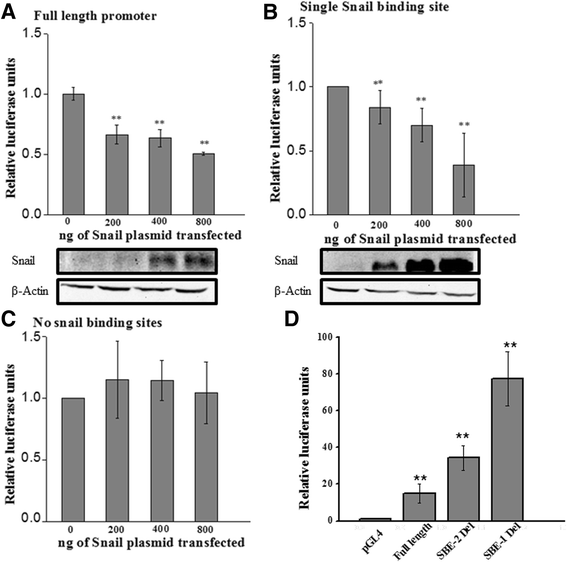
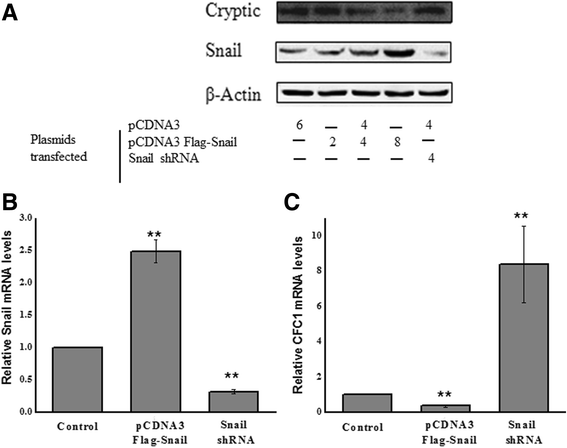
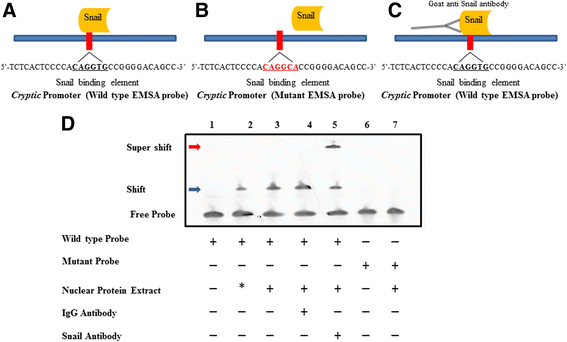
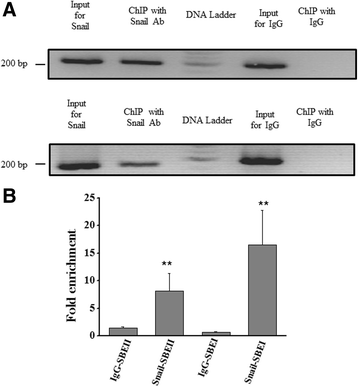
Results: Using promoter-analyses tools, we found strong evidence that the developmentally essential transcription factor Snail binds to the human Cryptic-promoter. We cloned the promoter-region of human Cryptic in a reporter gene and observed decreased Cryptic-promoter activation upon increasing Snail expression. Further, the expression of Cryptic is down-regulated upon exogenous Snail expression, validating the reporter assays and the previously identified role of Snail as a transcriptional repressor. Finally, we demonstrate using gel-shift assay that Snail in nuclear extract of PANC1 cells interacts with the promoter-construct bearing putative Snail binding sites and confirm this finding using chromatin immunoprecipitation assay.
Conclusions: Snail represses the expression of human Cryptic and therefore, might affect the signaling via Nodal that has previously been demonstrated to specify the left-right axis using the EGF-CFC co-receptors.
Keywords: Cryptic; EGF-CFC; Left-right-axis; Snail; Transcription.
Figures
Similar articles
-
A role of the cryptic gene in the correct establishment of the left-right axis.Curr Biol. 1999 Nov 18;9(22):1339-42. doi: 10.1016/s0960-9822(00)80059-7. Curr Biol. 1999. PMID: 10574770
-
Snail represses the expression of human phospholipid scramblase 4 gene.Gene. 2016 Oct 15;591(2):433-41. doi: 10.1016/j.gene.2016.06.050. Epub 2016 Jun 27. Gene. 2016. PMID: 27363667
-
XCR2, one of three Xenopus EGF-CFC genes, has a distinct role in the regulation of left-right patterning.Development. 2006 Jan;133(2):237-50. doi: 10.1242/dev.02188. Epub 2005 Dec 8. Development. 2006. PMID: 16339189
-
Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects.Nat Genet. 2000 Nov;26(3):365-9. doi: 10.1038/81695. Nat Genet. 2000. PMID: 11062482
-
The EGF-CFC gene family in vertebrate development.Trends Genet. 2000 Jul;16(7):303-9. doi: 10.1016/s0168-9525(00)02006-0. Trends Genet. 2000. PMID: 10858660 Review.
Cited by
-
Akap12beta supports asymmetric heart development via modulating the Kupffer's vesicle formation in zebrafish.BMB Rep. 2019 Aug;52(8):525-530. doi: 10.5483/BMBRep.2019.52.8.111. BMB Rep. 2019. PMID: 31383248 Free PMC article.
-
New Insights Into Breast and Chest Wall Asymmetry in the Aesthetic Patient.Aesthet Surg J Open Forum. 2024 Sep 14;6:ojae080. doi: 10.1093/asjof/ojae080. eCollection 2024. Aesthet Surg J Open Forum. 2024. PMID: 39670218 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

