Integrating a prospective pilot trial and patient-derived xenografts to trace metabolic changes associated with acute myeloid leukemia
- PMID: 27793157
- PMCID: PMC5086061
- DOI: 10.1186/s13045-016-0346-2
Integrating a prospective pilot trial and patient-derived xenografts to trace metabolic changes associated with acute myeloid leukemia
Abstract
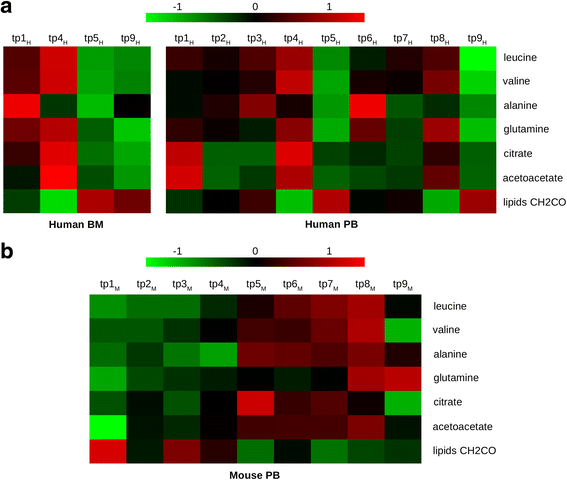
Despite the considerable progress in understanding the molecular bases of acute myeloid leukemia (AML), new tools to link disease biology to the unpredictable patient clinical course are still needed. Herein, high-throughput metabolomics, combined with the other "-omics" disciplines, holds promise in identifying disease-specific and clinically relevant features.In this study, we took advantage of nuclear magnetic resonance (NMR) to trace AML-associated metabolic trajectory employing two complementary strategies. On the one hand, we performed a prospective observational clinical trial to identify metabolic changes associated with blast clearance during the first two cycles of intensive chemotherapy in nine adult patients. On the other hand, to reduce the intrinsic variability associated with human samples and AML genetic heterogeneity, we analyzed the metabolic changes in the plasma of immunocompromised mice upon engraftment of primary human AML blasts.Combining the two longitudinal approaches, we narrowed our screen to seven common metabolites, for which we observed a mirror-like trajectory in mice and humans, tracing AML progression and remission, respectively. We interpreted this set of metabolites as a dynamic fingerprint of AML evolution.Overall, these NMR-based metabolomic data, to be consolidated in larger cohorts and integrated in more comprehensive system biology approaches, hold promise for providing valuable and non-redundant information on the systemic effects of leukemia.
Keywords: Acute myeloid leukemia; Metabolomics; Nuclear magnetic resonance; Patient-derived xenografts.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

