Bacteria as Emerging Indicators of Soil Condition
- PMID: 27793827
- PMCID: PMC5165110
- DOI: 10.1128/AEM.02826-16
Bacteria as Emerging Indicators of Soil Condition
Abstract
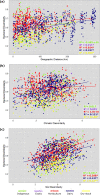
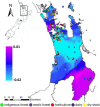
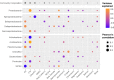
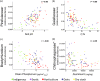
Bacterial communities are important for the health and productivity of soil ecosystems and have great potential as novel indicators of environmental perturbations. To assess how they are affected by anthropogenic activity and to determine their ability to provide alternative metrics of environmental health, we sought to define which soil variables bacteria respond to across multiple soil types and land uses. We determined, through 16S rRNA gene amplicon sequencing, the composition of bacterial communities in soil samples from 110 natural or human-impacted sites, located up to 300 km apart. Overall, soil bacterial communities varied more in response to changing soil environments than in response to changes in climate or increasing geographic distance. We identified strong correlations between the relative abundances of members of Pirellulaceae and soil pH, members of Gaiellaceae and carbon-to-nitrogen ratios, members of Bradyrhizobium and the levels of Olsen P (a measure of plant available phosphorus), and members of Chitinophagaceae and aluminum concentrations. These relationships between specific soil attributes and individual soil taxa not only highlight ecological characteristics of these organisms but also demonstrate the ability of key bacterial taxonomic groups to reflect the impact of specific anthropogenic activities, even in comparisons of samples across large geographic areas and diverse soil types. Overall, we provide strong evidence that there is scope to use relative taxon abundances as biological indicators of soil condition.
Importance: The impact of land use change and management on soil microbial community composition remains poorly understood. Therefore, we explored the relationship between a wide range of soil factors and soil bacterial community composition. We included variables related to anthropogenic activity and collected samples across a large spatial scale to interrogate the complex relationships between various bacterial community attributes and soil condition. We provide evidence of strong relationships between individual taxa and specific soil attributes even across large spatial scales and soil and land use types. Collectively, we were able to demonstrate the largely untapped potential of microorganisms to indicate the condition of soil and thereby influence the way that we monitor the effects of anthropogenic activity on soil ecosystems into the future.
Keywords: biogeography; biological indicator; soil health; soil microbiology.
Copyright © 2016 American Society for Microbiology.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

