Visualization of DNA G-quadruplexes in herpes simplex virus 1-infected cells
- PMID: 27794039
- PMCID: PMC5137459
- DOI: 10.1093/nar/gkw968
Visualization of DNA G-quadruplexes in herpes simplex virus 1-infected cells
Abstract
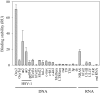
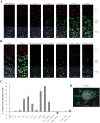
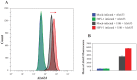
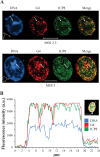
We have previously shown that clusters of guanine quadruplex (G4) structures can form in the human herpes simplex-1 (HSV-1) genome. Here we used immunofluorescence and immune-electron microscopy with a G4-specific monoclonal antibody to visualize G4 structures in HSV-1 infected cells. We found that G4 formation and localization within the cells was virus cycle dependent: viral G4s peaked at the time of viral DNA replication in the cell nucleus, moved to the nuclear membrane at the time of virus nuclear egress and were later found in HSV-1 immature virions released from the cell nucleus. Colocalization of G4s with ICP8, a viral DNA processing protein, was observed in viral replication compartments. G4s were lost upon treatment with DNAse and inhibitors of HSV-1 DNA replication. The notable increase in G4s upon HSV-1 infection suggests a key role of these structures in the HSV-1 biology and indicates new targets to control both the lytic and latent infection.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Huppert J.L. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008;37:1375–1384. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

