PHD2 Is a Regulator for Glycolytic Reprogramming in Macrophages
- PMID: 27795296
- PMCID: PMC5192080
- DOI: 10.1128/MCB.00236-16
PHD2 Is a Regulator for Glycolytic Reprogramming in Macrophages
Abstract
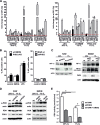
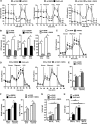
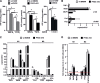
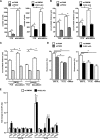
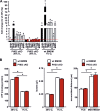
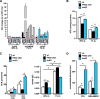
The prolyl-4-hydroxylase domain (PHD) enzymes are regarded as the molecular oxygen sensors. There is an interplay between oxygen availability and cellular metabolism, which in turn has significant effects on the functionality of innate immune cells, such as macrophages. However, if and how PHD enzymes affect macrophage metabolism are enigmatic. We hypothesized that macrophage metabolism and function can be controlled via manipulation of PHD2. We characterized the metabolic phenotypes of PHD2-deficient RAW cells and primary PHD2 knockout bone marrow-derived macrophages (BMDM). Both showed typical features of anaerobic glycolysis, which were paralleled by increased pyruvate dehydrogenase kinase 1 (PDK1) protein levels and a decreased pyruvate dehydrogenase enzyme activity. Metabolic alterations were associated with an impaired cellular functionality. Inhibition of PDK1 or knockout of hypoxia-inducible factor 1α (HIF-1α) reversed the metabolic phenotype and impaired the functionality of the PHD2-deficient RAW cells and BMDM. Taking these results together, we identified a critical role of PHD2 for a reversible glycolytic reprogramming in macrophages with a direct impact on their function. We suggest that PHD2 serves as an adjustable switch to control macrophage behavior.
Keywords: PDK; dioxygenases; hypoxia; macrophages; prolyl-4-hydroxylase domain.
Copyright © 2016 American Society for Microbiology.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
