Loss-of-function genetic tools for animal models: cross-species and cross-platform differences
- PMID: 27795562
- PMCID: PMC5206767
- DOI: 10.1038/nrg.2016.118
Loss-of-function genetic tools for animal models: cross-species and cross-platform differences
Abstract
Our understanding of the genetic mechanisms that underlie biological processes has relied extensively on loss-of-function (LOF) analyses. LOF methods target DNA, RNA or protein to reduce or to ablate gene function. By analysing the phenotypes that are caused by these perturbations the wild-type function of genes can be elucidated. Although all LOF methods reduce gene activity, the choice of approach (for example, mutagenesis, CRISPR-based gene editing, RNA interference, morpholinos or pharmacological inhibition) can have a major effect on phenotypic outcomes. Interpretation of the LOF phenotype must take into account the biological process that is targeted by each method. The practicality and efficiency of LOF methods also vary considerably between model systems. We describe parameters for choosing the optimal combination of method and system, and for interpreting phenotypes within the constraints of each method.
Figures
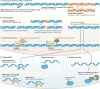
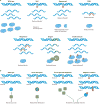
References
-
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. - PubMed
-
- Haffter P, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. - PubMed
-
- Driever W, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. References 4 and 5 are leading papers that describe the first large-scale ENU screens conducted in zebrafish. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

