Functional Characterization of Inflammatory Bowel Disease-Associated Gut Dysbiosis in Gnotobiotic Mice
- PMID: 27795980
- PMCID: PMC5042563
- DOI: 10.1016/j.jcmgh.2016.02.003
Functional Characterization of Inflammatory Bowel Disease-Associated Gut Dysbiosis in Gnotobiotic Mice
Abstract
Background & aims: Gut dysbiosis is closely involved in the pathogenesis of inflammatory bowel disease (IBD). However, it remains unclear whether IBD-associated gut dysbiosis contributes to disease pathogenesis or is merely secondary to intestinal inflammation. We established a humanized gnotobiotic (hGB) mouse system to assess the functional role of gut dysbiosis associated with 2 types of IBD: Crohn's disease (CD) and ulcerative colitis (UC).
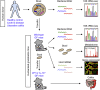
Methods: Germ-free mice were colonized by the gut microbiota isolated from patients with CD and UC, and healthy controls. Microbiome analysis, bacterial functional gene analysis, luminal metabolome analysis, and host gene expression analysis were performed in hGB mice. Moreover, the colitogenic capacity of IBD-associated microbiota was evaluated by colonizing germ-free colitis-prone interleukin 10-deficient mice with dysbiotic patients' microbiota.
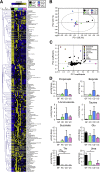
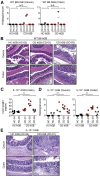
Results: Although the microbial composition seen in donor patients' microbiota was not completely reproduced in hGB mice, some dysbiotic features of the CD and UC microbiota (eg, decreased diversity, alteration of bacterial metabolic functions) were recapitulated in hGB mice, suggesting that microbial community alterations, characteristic for IBD, can be reproduced in hGB mice. In addition, colonization by the IBD-associated microbiota induced a proinflammatory gene expression profile in the gut that resembles the immunologic signatures found in CD patients. Furthermore, CD microbiota triggered more severe colitis than healthy control microbiota when colonized in germ-free interleukin 10-deficient mice.
Conclusions: Dysbiosis potentially contributes to the pathogenesis of IBD by augmenting host proinflammatory immune responses. Transcript profiling: GSE73882.
Keywords: CD, Crohn's disease; CE-TOFMS, capillary electrophoresis time-of-flight mass spectrometry; Crohn's Disease; Dysbiosis; GB, gnotobiotic; GF, germ-free; IBD, inflammatory bowel disease; IFN, interferon; IL, interleukin; ILC, innate lymphoid cell; IVC, individual ventilated cage; Microbiota; NK, natural killer; OTU, operational taxonomic unit; SCFA, short-chain fatty acid; Th, T helper; UC, ulcerative colitis; Ulcerative Colitis; WT, wild type; hGB, humanized gnotobiotic; rRNA, ribosomal RNA.
Conflict of interest statement
The authors declared no conflict of interest.
Figures



References
-
- Kamada N., Seo S.U., Chen G.Y. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. - PubMed
-
- Sartor R.B. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. - PubMed
-
- Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

