KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies
- PMID: 27797770
- PMCID: PMC5408915
- DOI: 10.1093/bioinformatics/btw663
KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies
Abstract
Motivation: De novo assembly of whole genome shotgun (WGS) next-generation sequencing (NGS) data benefits from high-quality input with high coverage. However, in practice, determining the quality and quantity of useful reads quickly and in a reference-free manner is not trivial. Gaining a better understanding of the WGS data, and how that data is utilized by assemblers, provides useful insights that can inform the assembly process and result in better assemblies.
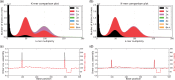
Results: We present the K-mer Analysis Toolkit (KAT): a multi-purpose software toolkit for reference-free quality control (QC) of WGS reads and de novo genome assemblies, primarily via their k-mer frequencies and GC composition. KAT enables users to assess levels of errors, bias and contamination at various stages of the assembly process. In this paper we highlight KAT's ability to provide valuable insights into assembly composition and quality of genome assemblies through pairwise comparison of k-mers present in both input reads and the assemblies.
Availability and implementation: KAT is available under the GPLv3 license at: https://github.com/TGAC/KAT .
Contact: bernardo.clavijo@earlham.ac.uk.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2016. Published by Oxford University Press.
Figures
References
-
- Metzker M.L. (2010) Sequencing technologies - the next generation. Nat. Rev. Genet., 11, 31–46. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

