Genetically Targeted All-Optical Electrophysiology with a Transgenic Cre-Dependent Optopatch Mouse
- PMID: 27798186
- PMCID: PMC5098841
- DOI: 10.1523/JNEUROSCI.1582-16.2016
Genetically Targeted All-Optical Electrophysiology with a Transgenic Cre-Dependent Optopatch Mouse
Abstract
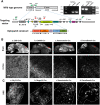
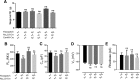
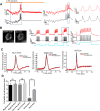
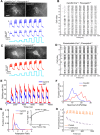
Recent advances in optogenetics have enabled simultaneous optical perturbation and optical readout of membrane potential in diverse cell types. Here, we develop and characterize a Cre-dependent transgenic Optopatch2 mouse line that we call Floxopatch. The animals expressed a blue-shifted channelrhodopsin, CheRiff, and a near infrared Archaerhodopsin-derived voltage indicator, QuasAr2, via targeted knock-in at the rosa26 locus. In Optopatch-expressing animals, we tested for overall health, genetically targeted expression, and function of the optogenetic components. In offspring of Floxopatch mice crossed with a variety of Cre driver lines, we observed spontaneous and optically evoked activity in vitro in acute brain slices and in vivo in somatosensory ganglia. Cell-type-specific expression allowed classification and characterization of neuronal subtypes based on their firing patterns. The Floxopatch mouse line is a useful tool for fast and sensitive characterization of neural activity in genetically specified cell types in intact tissue.
Significance statement: Optical recordings of neural activity offer the promise of rapid and spatially resolved mapping of neural function. Calcium imaging has been widely applied in this mode, but is insensitive to the details of action potential waveforms and subthreshold events. Simultaneous optical perturbation and optical readout of single-cell electrical activity ("Optopatch") has been demonstrated in cultured neurons and in organotypic brain slices, but not in acute brain slices or in vivo Here, we describe a transgenic mouse in which expression of Optopatch constructs is controlled by the Cre-recombinase enzyme. This animal enables fast and robust optical measurements of single-cell electrical excitability in acute brain slices and in somatosensory ganglia in vivo, opening the door to rapid optical mapping of neuronal excitability.
Keywords: optogenetics; optopatch; transgenic mice; voltage imaging.
Copyright © 2016 the authors 0270-6474/16/3611059-15$15.00/0.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
