Multiple dynamin family members collaborate to drive mitochondrial division
- PMID: 27798601
- PMCID: PMC5656044
- DOI: 10.1038/nature20555
Multiple dynamin family members collaborate to drive mitochondrial division
Abstract
Mitochondria cannot be generated de novo; they must grow, replicate their genome, and divide in order to be inherited by each daughter cell during mitosis. Mitochondrial division is a structural challenge that requires the substantial remodelling of membrane morphology. Although division factors differ across organisms, the need for multiple constriction steps and a dynamin-related protein (Drp1, Dnm1 in yeast) has been conserved. In mammalian cells, mitochondrial division has been shown to proceed with at least two sequential constriction steps: the endoplasmic reticulum and actin must first collaborate to generate constrictions suitable for Drp1 assembly on the mitochondrial outer membrane; Drp1 then further constricts membranes until mitochondrial fission occurs. In vitro experiments, however, indicate that Drp1 does not have the dynamic range to complete membrane fission. In contrast to Drp1, the neuron-specific classical dynamin dynamin-1 (Dyn1) has been shown to assemble on narrower lipid profiles and facilitate spontaneous membrane fission upon GTP hydrolysis. Here we report that the ubiquitously expressed classical dynamin-2 (Dyn2) is a fundamental component of the mitochondrial division machinery. A combination of live-cell and electron microscopy in three different mammalian cell lines reveals that Dyn2 works in concert with Drp1 to orchestrate sequential constriction events that build up to division. Our work underscores the biophysical limitations of Drp1 and positions Dyn2, which has intrinsic membrane fission properties, at the final step of mitochondrial division.
Figures
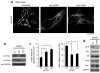

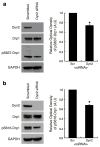
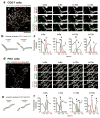

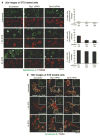

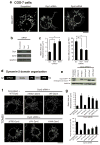

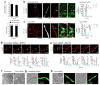
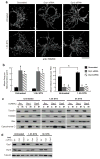
Comment in
-
Cell biology: Double agents for mitochondrial division.Nature. 2016 Dec 1;540(7631):43-44. doi: 10.1038/nature20482. Epub 2016 Nov 23. Nature. 2016. PMID: 27880759 No abstract available.
-
Mitochondrial fission requires DRP1 but not dynamins.Nature. 2019 Jun;570(7761):E34-E42. doi: 10.1038/s41586-019-1296-y. Epub 2019 Jun 19. Nature. 2019. PMID: 31217603 No abstract available.
References
-
- Goldstein S, Moerman EJ, Porter K. High-voltage electron microscopy of human diploid fibroblasts during ageing in vitro. Morphometric analysis of mitochondria. Exp Cell Res. 1984;154:101–11. - PubMed
-
- Kim HW, et al. Efficient and accurate analysis of mitochondrial morphology in a whole cell with a high-voltage electron microscopy. J Electron Microsc (Tokyo) 2012;61:127–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

