Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours
- PMID: 27798604
- PMCID: PMC5812439
- DOI: 10.1038/ncb3434
Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours
Abstract
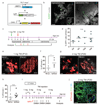
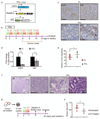
The mammary gland is composed of a complex cellular hierarchy with unusual postnatal plasticity. The identities of stem/progenitor cell populations, as well as tumour-initiating cells that give rise to breast cancer, are incompletely understood. Here we show that Lgr6 marks rare populations of cells in both basal and luminal mammary gland compartments in mice. Lineage tracing analysis showed that Lgr6+ cells are unipotent progenitors, which expand clonally during puberty but diminish in adulthood. In pregnancy or following stimulation with ovarian hormones, adult Lgr6+ cells regained proliferative potency and their progeny formed alveoli over repeated pregnancies. Oncogenic mutations in Lgr6+ cells resulted in expansion of luminal cells, culminating in mammary gland tumours. Conversely, depletion of Lgr6+ cells in the MMTV-PyMT model of mammary tumorigenesis significantly impaired tumour growth. Thus, Lgr6 marks mammary gland progenitor cells that can initiate tumours, and cells of luminal breast tumours required for efficient tumour maintenance.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






References
-
- Inman JL, Robertson C, Mott JD, Bissell MJ. Mammary gland development: cell fate specification, stem cells and the microenvironment. Development. 2015;142:1028–1042. - PubMed
-
- Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. - PubMed
-
- Wang D, et al. Identification of multipotent mammary stem cells by protein C receptor expression. Nature. 2015;517:81–84. - PubMed
-
- van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

