Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages
- PMID: 27798628
- PMCID: PMC5238942
- DOI: 10.1038/ng.3704
Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages
Abstract
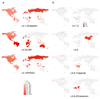
Generalist and specialist species differ in the breadth of their ecological niches. Little is known about the niche width of obligate human pathogens. Here we analyzed a global collection of Mycobacterium tuberculosis lineage 4 clinical isolates, the most geographically widespread cause of human tuberculosis. We show that lineage 4 comprises globally distributed and geographically restricted sublineages, suggesting a distinction between generalists and specialists. Population genomic analyses showed that, whereas the majority of human T cell epitopes were conserved in all sublineages, the proportion of variable epitopes was higher in generalists. Our data further support a European origin for the most common generalist sublineage. Hence, the global success of lineage 4 reflects distinct strategies adopted by different sublineages and the influence of human migration.
Conflict of interest statement
Statement The authors have no competing interests as defined by Springer Nature, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Figures



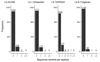

Comment in
-
Human host range of Mycobacterium tuberculosis.Nat Genet. 2016 Nov 29;48(12):1453-1454. doi: 10.1038/ng.3724. Nat Genet. 2016. PMID: 27898082
References
-
- Futuyma DJ, Moreno G. The Evolution of Ecological Specialization. Annual Review of Ecology and Systematics. 1988;19:207–233.
-
- Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–77. - PubMed
-
- Woolhouse ME, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–12. - PubMed
-
- Kirzinger MW, Stavrinides J. Host specificity determinants as a genetic continuum. Trends Microbiol. 2012;20:88–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

