Application of High-Throughput Next-Generation Sequencing for HLA Typing on Buccal Extracted DNA: Results from over 10,000 Donor Recruitment Samples
- PMID: 27798706
- PMCID: PMC5087893
- DOI: 10.1371/journal.pone.0165810
Application of High-Throughput Next-Generation Sequencing for HLA Typing on Buccal Extracted DNA: Results from over 10,000 Donor Recruitment Samples
Abstract
Background: Unambiguous HLA typing is important in hematopoietic stem cell transplantation (HSCT), HLA disease association studies, and solid organ transplantation. However, current molecular typing methods only interrogate the antigen recognition site (ARS) of HLA genes, resulting in many cis-trans ambiguities that require additional typing methods to resolve. Here we report high-resolution HLA typing of 10,063 National Marrow Donor Program (NMDP) registry donors using long-range PCR by next generation sequencing (NGS) approach on buccal swab DNA.
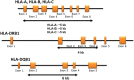
Methods: Multiplex long-range PCR primers amplified the full-length of HLA class I genes (A, B, C) from promotor to 3' UTR. Class II genes (DRB1, DQB1) were amplified from exon 2 through part of exon 4. PCR amplicons were pooled and sheared using Covaris fragmentation. Library preparation was performed using the Illumina TruSeq Nano kit on the Beckman FX automated platform. Each sample was tagged with a unique barcode, followed by 2×250 bp paired-end sequencing on the Illumina MiSeq. HLA typing was assigned using Omixon Twin software that combines two independent computational algorithms to ensure high confidence in allele calling. Consensus sequence and typing results were reported in Histoimmunogenetics Markup Language (HML) format. All homozygous alleles were confirmed by Luminex SSO typing and exon novelties were confirmed by Sanger sequencing.
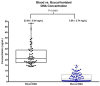
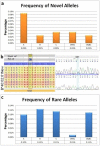
Results: Using this automated workflow, over 10,063 NMDP registry donors were successfully typed under high-resolution by NGS. Despite known challenges of nucleic acid degradation and low DNA concentration commonly associated with buccal-based specimens, 97.8% of samples were successfully amplified using long-range PCR. Among these, 98.2% were successfully reported by NGS, with an accuracy rate of 99.84% in an independent blind Quality Control audit performed by the NDMP. In this study, NGS-HLA typing identified 23 null alleles (0.023%), 92 rare alleles (0.091%) and 42 exon novelties (0.042%).
Conclusion: Long-range, unambiguous HLA genotyping is achievable on clinical buccal swab-extracted DNA. Importantly, full-length gene sequencing and the ability to curate full sequence data will permit future interrogation of the impact of introns, expanded exons, and other gene regulatory sequences on clinical outcomes in transplantation.
Conflict of interest statement
Katsuyuki Saito is employed by One Lambda, Thermo Fisher Scientific. Tim Hague, Agnes Pasztor, Gyorgy Horvath and Krisztina Rigo are employed by Omixon LTD. There are no patents, products in development or marketed products to declare. This does not alter our adherence to all the PLOS ONE policies on sharing data and materials.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

