Post-transcriptional control of executioner caspases by RNA-binding proteins
- PMID: 27798844
- PMCID: PMC5088569
- DOI: 10.1101/gad.285726.116
Post-transcriptional control of executioner caspases by RNA-binding proteins
Abstract
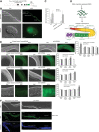
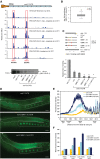
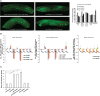
Caspases are key components of apoptotic pathways. Regulation of caspases occurs at several levels, including transcription, proteolytic processing, inhibition of enzymatic function, and protein degradation. In contrast, little is known about the extent of post-transcriptional control of caspases. Here, we describe four conserved RNA-binding proteins (RBPs)-PUF-8, MEX-3, GLD-1, and CGH-1-that sequentially repress the CED-3 caspase in distinct regions of the Caenorhabditis elegans germline. We demonstrate that GLD-1 represses ced-3 mRNA translation via two binding sites in its 3' untranslated region (UTR), thereby ensuring a dual control of unwanted cell death: at the level of p53/CEP-1 and at the executioner caspase level. Moreover, we identified seven RBPs that regulate human caspase-3 expression and/or activation, including human PUF-8, GLD-1, and CGH-1 homologs PUM1, QKI, and DDX6. Given the presence of unusually long executioner caspase 3' UTRs in many metazoans, translational control of executioner caspases by RBPs might be a strategy used widely across the animal kingdom to control apoptosis.
Keywords: CED-3; GLD-1; RNA-binding proteins; apoptosis; caspase; caspase-3; translational control.
© 2016 Subasic et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Similar articles
-
The RNA-binding proteins PUF-5, PUF-6, and PUF-7 reveal multiple systems for maternal mRNA regulation during C. elegans oogenesis.Dev Biol. 2007 Mar 15;303(2):635-49. doi: 10.1016/j.ydbio.2006.12.004. Epub 2006 Dec 8. Dev Biol. 2007. PMID: 17234175
-
Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis.Cell. 2005 Feb 11;120(3):357-68. doi: 10.1016/j.cell.2004.12.009. Cell. 2005. PMID: 15707894
-
Both the caspase CSP-1 and a caspase-independent pathway promote programmed cell death in parallel to the canonical pathway for apoptosis in Caenorhabditis elegans.PLoS Genet. 2013;9(3):e1003341. doi: 10.1371/journal.pgen.1003341. Epub 2013 Mar 7. PLoS Genet. 2013. PMID: 23505386 Free PMC article.
-
Apoptosis: a process with a (beta)NAC for complexity.Cell. 2003 Sep 19;114(6):659-61. doi: 10.1016/s0092-8674(03)00717-7. Cell. 2003. PMID: 14505566 Review.
-
Methods for detection and analysis of apoptosis signaling in the C. elegans germline.Methods. 2013 Jun 1;61(2):174-82. doi: 10.1016/j.ymeth.2013.04.022. Epub 2013 May 2. Methods. 2013. PMID: 23643851 Review.
Cited by
-
DeepCleave: a deep learning predictor for caspase and matrix metalloprotease substrates and cleavage sites.Bioinformatics. 2020 Feb 15;36(4):1057-1065. doi: 10.1093/bioinformatics/btz721. Bioinformatics. 2020. PMID: 31566664 Free PMC article.
-
Pumilio RNA-Binding Family Member 1 Plays a Promoting Role on Pancreatic Cancer Angiogenesis.Can J Gastroenterol Hepatol. 2022 Dec 14;2022:9202531. doi: 10.1155/2022/9202531. eCollection 2022. Can J Gastroenterol Hepatol. 2022. PMID: 39296516 Free PMC article.
-
PUF-8, a C. elegans ortholog of the RNA-binding proteins PUM1 and PUM2, is required for robustness of the cell death fate.Development. 2023 Oct 1;150(19):dev201167. doi: 10.1242/dev.201167. Epub 2023 Oct 6. Development. 2023. PMID: 37747106 Free PMC article.
-
MINA-1 and WAGO-4 are part of regulatory network coordinating germ cell death and RNAi in C. elegans.Cell Death Differ. 2019 Oct;26(10):2157-2178. doi: 10.1038/s41418-019-0291-z. Epub 2019 Feb 6. Cell Death Differ. 2019. PMID: 30728462 Free PMC article.
-
PUM1 knockdown prevents tumor progression by activating the PERK/eIF2/ATF4 signaling pathway in pancreatic adenocarcinoma cells.Cell Death Dis. 2019 Aug 8;10(8):595. doi: 10.1038/s41419-019-1839-z. Cell Death Dis. 2019. PMID: 31395860 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
