Protein stabilization improves STAT3 function in autosomal dominant hyper-IgE syndrome
- PMID: 27799162
- PMCID: PMC5201093
- DOI: 10.1182/blood-2016-02-702373
Protein stabilization improves STAT3 function in autosomal dominant hyper-IgE syndrome
Abstract
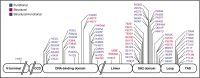
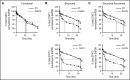
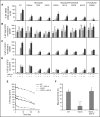
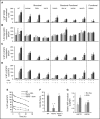
Autosomal dominant hyper-IgE syndrome (AD-HIES) is caused by dominant-negative mutations in STAT3; however, the molecular basis for mutant STAT3 allele dysfunction is unclear and treatment remains supportive. We hypothesized that AD-HIES mutations decrease STAT3 protein stability and that mutant STAT3 activity can be improved by agents that increase chaperone protein activity. We used computer modeling to characterize the effect of STAT3 mutations on protein stability. We measured STAT3 protein half-life (t1/2) and determined levels of STAT3 phosphorylated on tyrosine (Y) 705 (pY-STAT3) and mRNA levels of STAT3 gene targets in Epstein-Barr virus-transformed B (EBV) cells, human peripheral blood mononuclear cells (PBMCs), and mouse splenocytes incubated without or with chaperone protein modulators-HSF1A, a small-molecule TRiC modulator, or geranylgeranylacetone (GGA), a drug that upregulates heat shock protein (HSP) 70 and HSP90. Computer modeling predicted that 81% of AD-HIES mutations are destabilizing. STAT3 protein t1/2 in EBV cells from AD-HIES patients with destabilizing STAT3 mutations was markedly reduced. Treatment of EBV cells containing destabilizing STAT3 mutations with either HSF1A or GGA normalized STAT3 t1/2, increased pY-STAT3 levels, and increased mRNA levels of STAT3 target genes up to 79% of control. In addition, treatment of human PBMCs or mouse splenocytes containing destabilizing STAT3 mutations with either HSF1A or GGA increased levels of cytokine-activated pY-STAT3 within human CD4+ and CD8+ T cells and numbers of IL-17-producing CD4+ mouse splenocytes, respectively. Thus, most AD-HIES STAT3 mutations are destabilizing; agents that modulate chaperone protein function improve STAT3 stability and activity in T cells and may provide a specific treatment.
Figures

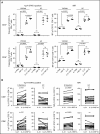
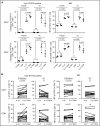
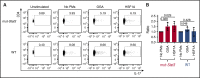
Comment in
-
When half a glass of STAT3 is just not enough.Blood. 2016 Dec 29;128(26):3020-3021. doi: 10.1182/blood-2016-11-750539. Blood. 2016. PMID: 28034865 No abstract available.
References
-
- Davis SD, Schaller J, Wedgwood RJ. Job’s Syndrome. Recurrent, “cold”, staphylococcal abscesses. Lancet. 1966;1(7445):1013-1015. - PubMed
-
- Holland SM, DeLeo FR, Elloumi HZ, et al. . STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357(16):1608-1619. - PubMed
-
- Jiao H, Tóth B, Erdos M, et al. . Novel and recurrent STAT3 mutations in hyper-IgE syndrome patients from different ethnic groups. Mol Immunol. 2008;46(1):202-206. - PubMed
-
- Minegishi Y, Saito M, Tsuchiya S, et al. . Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448(7157):1058-1062. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

