MutS2 Promotes Homologous Recombination in Bacillus subtilis
- PMID: 27799325
- PMCID: PMC5198493
- DOI: 10.1128/JB.00682-16
MutS2 Promotes Homologous Recombination in Bacillus subtilis
Abstract
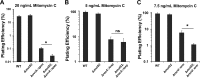
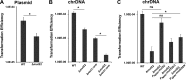
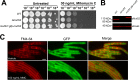
Bacterial MutS proteins are subdivided into two families, MutS1 and MutS2. MutS1 family members recognize DNA replication errors during their participation in the well-characterized mismatch repair (MMR) pathway. In contrast to the well-described function of MutS1, the function of MutS2 in bacteria has remained less clear. In Helicobacter pylori and Thermus thermophilus, MutS2 has been shown to suppress homologous recombination. The role of MutS2 is unknown in the Gram-positive bacterium Bacillus subtilis In this work, we investigated the contribution of MutS2 to maintaining genome integrity in B. subtilis We found that deletion of mutS2 renders B. subtilis sensitive to the natural antibiotic mitomycin C (MMC), which requires homologous recombination for repair. We demonstrate that the C-terminal small MutS-related (Smr) domain is necessary but not sufficient for tolerance to MMC. Further, we developed a CRISPR/Cas9 genome editing system to test if the inducible prophage PBSX was the underlying cause of the observed MMC sensitivity. Genetic analysis revealed that MMC sensitivity was dependent on recombination and not on nucleotide excision repair or a symptom of prophage PBSX replication and cell lysis. We found that deletion of mutS2 resulted in decreased transformation efficiency using both plasmid and chromosomal DNA. Further, deletion of mutS2 in a strain lacking the Holliday junction endonuclease gene recU resulted in increased MMC sensitivity and decreased transformation efficiency, suggesting that MutS2 could function redundantly with RecU. Together, our results support a model where B. subtilis MutS2 helps to promote homologous recombination, demonstrating a new function for bacterial MutS2.
Importance: Cells contain pathways that promote or inhibit recombination. MutS2 homologs are Smr-endonuclease domain-containing proteins that have been shown to function in antirecombination in some bacteria. We present evidence that B. subtilis MutS2 promotes recombination, providing a new function for MutS2. We found that cells lacking mutS2 are sensitive to DNA damage that requires homologous recombination for repair and have reduced transformation efficiency. Further analysis indicates that the C-terminal Smr domain requires the N-terminal portion of MutS2 for function in vivo Moreover, we show that a mutS2 deletion is additive with a recU deletion, suggesting that these proteins have a redundant function in homologous recombination. Together, our study shows that MutS2 proteins have adapted different functions that impact recombination.
Keywords: CRISPR; DNA damage; DNA recombination; MutS.
Copyright © 2016 American Society for Microbiology.
Figures



References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. 2006. DNA repair and mutagenesis, 2nd ed American Society for Microbiology, Washington, DC.
-
- Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, d'Adda di Fagagna F, Devine KM, Bowater RP, Jeggo PA, Jackson SP, Doherty AJ. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297:1686–1689. doi:10.1126/science.1074584. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

