Using Next-Generation Sequencing for DNA Barcoding: Capturing Allelic Variation in ITS2
- PMID: 27799340
- PMCID: PMC5217108
- DOI: 10.1534/g3.116.036145
Using Next-Generation Sequencing for DNA Barcoding: Capturing Allelic Variation in ITS2
Abstract
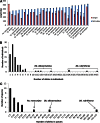
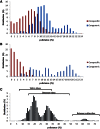
Internal Transcribed Spacer 2 (ITS2) is a popular DNA barcoding marker; however, in some animal species it is hypervariable and therefore difficult to sequence with traditional methods. With next-generation sequencing (NGS) it is possible to sequence all gene variants despite the presence of single nucleotide polymorphisms (SNPs), insertions/deletions (indels), homopolymeric regions, and microsatellites. Our aim was to compare the performance of Sanger sequencing and NGS amplicon sequencing in characterizing ITS2 in 26 mosquito species represented by 88 samples. The suitability of ITS2 as a DNA barcoding marker for mosquitoes, and its allelic diversity in individuals and species, was also assessed. Compared to Sanger sequencing, NGS was able to characterize the ITS2 region to a greater extent, with resolution within and between individuals and species that was previously not possible. A total of 382 unique sequences (alleles) were generated from the 88 mosquito specimens, demonstrating the diversity present that has been overlooked by traditional sequencing methods. Multiple indels and microsatellites were present in the ITS2 alleles, which were often specific to species or genera, causing variation in sequence length. As a barcoding marker, ITS2 was able to separate all of the species, apart from members of the Culex pipiens complex, providing the same resolution as the commonly used Cytochrome Oxidase I (COI). The ability to cost-effectively sequence hypervariable markers makes NGS an invaluable tool with many applications in the DNA barcoding field, and provides insights into the limitations of previous studies and techniques.
Keywords: Culicidae; NGS; amplicon sequencing; indels; microsatellites.
Copyright © 2017 Batovska et al.
Figures



References
-
- Álvarez I., Wendel J. F., 2003. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29: 417–434. - PubMed
-
- Aspen S., Crabtree M. B., Savage H. M., 2003. Polymerase chain reaction assay identifies Culex nigripalpus: part of an assay for molecular identification of the common Culex (Culex) mosquitoes of the eastern United States. J. Am. Mosq. Control Assoc. 19: 115–120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
