IMG/VR: a database of cultured and uncultured DNA Viruses and retroviruses
- PMID: 27799466
- PMCID: PMC5210529
- DOI: 10.1093/nar/gkw1030
IMG/VR: a database of cultured and uncultured DNA Viruses and retroviruses
Abstract
Viruses represent the most abundant life forms on the planet. Recent experimental and computational improvements have led to a dramatic increase in the number of viral genome sequences identified primarily from metagenomic samples. As a result of the expanding catalog of metagenomic viral sequences, there exists a need for a comprehensive computational platform integrating all these sequences with associated metadata and analytical tools. Here we present IMG/VR (https://img.jgi.doe.gov/vr/), the largest publicly available database of 3908 isolate reference DNA viruses with 264 413 computationally identified viral contigs from >6000 ecologically diverse metagenomic samples. Approximately half of the viral contigs are grouped into genetically distinct quasi-species clusters. Microbial hosts are predicted for 20 000 viral sequences, revealing nine microbial phyla previously unreported to be infected by viruses. Viral sequences can be queried using a variety of associated metadata, including habitat type and geographic location of the samples, or taxonomic classification according to hallmark viral genes. IMG/VR has a user-friendly interface that allows users to interrogate all integrated data and interact by comparing with external sequences, thus serving as an essential resource in the viral genomics community.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
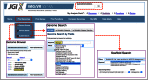
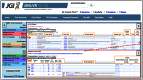
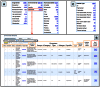

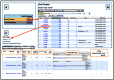

References
-
- Suttle C.A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. - PubMed
-
- Gomez P., Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–109. - PubMed
-
- Pal C., Macia M.D., Oliver A., Schachar I., Buckling A. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature. 2007;450:1079–1081. - PubMed
-
- Brum J.R., Sullivan M.B. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 2015;13:147–159. - PubMed
-
- Fuhrman J.A. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

