pMHC affinity controls duration of CD8+ T cell-DC interactions and imprints timing of effector differentiation versus expansion
- PMID: 27799622
- PMCID: PMC5110015
- DOI: 10.1084/jem.20160206
pMHC affinity controls duration of CD8+ T cell-DC interactions and imprints timing of effector differentiation versus expansion
Abstract
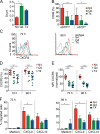
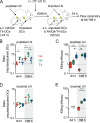
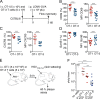
During adaptive immune responses, CD8+ T cells with low TCR affinities are released early into the circulation before high-affinity clones become dominant at later time points. How functional avidity maturation is orchestrated in lymphoid tissue and how low-affinity cells contribute to host protection remains unclear. In this study, we used intravital imaging of reactive lymph nodes (LNs) to show that T cells rapidly attached to dendritic cells irrespective of TCR affinity, whereas one day later, the duration of these stable interactions ceased progressively with lowering peptide major histocompatibility complex (pMHC) affinity. This correlated inversely BATF (basic leucine zipper transcription factor, ATF-like) and IRF4 (interferon-regulated factor 4) induction and timing of effector differentiation, as low affinity-primed T cells acquired cytotoxic activity earlier than high affinity-primed ones. After activation, low-affinity effector CD8+ T cells accumulated at efferent lymphatic vessels for egress, whereas high affinity-stimulated CD8+ T cells moved to interfollicular regions in a CXCR3-dependent manner for sustained pMHC stimulation and prolonged expansion. The early release of low-affinity effector T cells led to rapid target cell elimination outside reactive LNs. Our data provide a model for affinity-dependent spatiotemporal orchestration of CD8+ T cell activation inside LNs leading to functional avidity maturation and uncover a role for low-affinity effector T cells during early microbial containment.
© 2016 Ozga et al.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

