Tunable Helicity, Stability and DNA-Binding Properties of Short Peptides with Hybrid Metal Coordination Motifs
- PMID: 27800151
- PMCID: PMC5085262
- DOI: 10.1039/C6SC00826G
Tunable Helicity, Stability and DNA-Binding Properties of Short Peptides with Hybrid Metal Coordination Motifs
Abstract
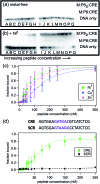
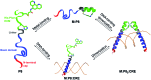
Given the prevalent role of α-helical motifs on protein surfaces in mediating protein-protein and protein-DNA interactions, there have been significant efforts to develop strategies to induce α-helicity in short, unstructured peptides to interrogate such interactions. Toward this goal, we have recently introduced hybrid metal coordination motifs (HCMs). HCMs combine a natural metal-binding amino acid side chain with a synthetic chelating group that are appropriately positioned in a peptide sequence to stabilize an α-helical conformation upon metal coordination. Here, we present a series of short peptides modified with HCMs consisting of a His and a phenanthroline group at i and i+7 positions that can induce α-helicity in a metal-tunable fashion as well as direct the formation of discrete dimeric architectures for recognition of biological targets. We show that the induction of α-helicity can be further modulated by secondary sphere interactions between amino acids at the i+4 position and the HCM. A frequently cited drawback of the use of peptides as therapeutics is their propensity to be quickly digested by proteases; here, we observe an enhancement of up to ∼100-fold in the half-lifes of the metal-bound HCM-peptides in the presence of trypsin. Finally, we show that an HCM-bearing peptide sequence, which contains the DNA-recognition domain of a bZIP protein but is devoid of the obligate dimerization domain, can dimerize with the proper geometry and in an α-helical conformation to bind a cognate DNA sequence with high affinities (Kd≥ 65 nM), again in a metal-tunable manner.
Figures
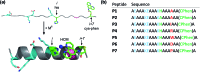




Similar articles
-
Functional, metal-based crosslinkers for α-helix induction in short peptides.Chem Sci. 2013 Sep;4(9):3740-3747. doi: 10.1039/C3SC50858G. Chem Sci. 2013. PMID: 24156013 Free PMC article.
-
Modular and versatile hybrid coordination motifs on alpha-helical protein surfaces.Inorg Chem. 2010 Aug 2;49(15):7106-15. doi: 10.1021/ic100926g. Inorg Chem. 2010. PMID: 20617830 Free PMC article.
-
The role of primary sequence in helical control compared across short α- and β(3)-peptides.J Biomol Struct Dyn. 2015;33(3):597-605. doi: 10.1080/07391102.2014.897260. Epub 2014 Mar 13. J Biomol Struct Dyn. 2015. PMID: 24625290
-
Controlled protein dimerization through hybrid coordination motifs.Inorg Chem. 2010 May 3;49(9):4362-9. doi: 10.1021/ic100534y. Inorg Chem. 2010. PMID: 20377257 Free PMC article.
-
Effect of chain length on the formation and stability of synthetic alpha-helical coiled coils.Biochemistry. 1994 Dec 27;33(51):15501-10. doi: 10.1021/bi00255a032. Biochemistry. 1994. PMID: 7803412
Cited by
-
Metal-Dependent DNA Recognition and Cell Internalization of Designed, Basic Peptides.J Am Chem Soc. 2017 Nov 15;139(45):16188-16193. doi: 10.1021/jacs.7b07422. Epub 2017 Nov 1. J Am Chem Soc. 2017. PMID: 29056048 Free PMC article.
-
Folding of unstructured peptoids and formation of hetero-bimetallic peptoid complexes upon side-chain-to-metal coordination.Chem Sci. 2018 Oct 17;10(2):620-632. doi: 10.1039/c8sc03616k. eCollection 2019 Jan 14. Chem Sci. 2018. PMID: 30713653 Free PMC article.
-
Activation of the cGAS-sting Pathway Mediated by Nanocomplexes for Tumor Therapy.Curr Pharm Des. 2025;31(20):1604-1618. doi: 10.2174/0113816128339788241221160639. Curr Pharm Des. 2025. PMID: 39819537 Review.
References
-
- Burkhard P., Stetefeld J., Strelkov S. V. Trends Cell Biol. 2001;11:82–88. - PubMed
-
- Lupas A. N. and Gruber M., in Advances in Protein Chemistry, Academic Press, 2005, vol. 70, pp. 37–38. - PubMed
-
- Youle R. J., Strasser A. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. - PubMed
-
- Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. Science. 1996;274:948–953. - PubMed
-
- Busch S. J., Sassone-Corsi P. Trends Genet. 1990;6:36–40. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

