Global Proteomics Analysis of Protein Lysine Methylation
- PMID: 27801517
- PMCID: PMC5222891
- DOI: 10.1002/cpps.16
Global Proteomics Analysis of Protein Lysine Methylation
Abstract
Lysine methylation is a common protein post-translational modification dynamically mediated by protein lysine methyltransferases (PKMTs) and protein lysine demethylases (PKDMs). Beyond histone proteins, lysine methylation on non-histone proteins plays a substantial role in a variety of functions in cells and is closely associated with diseases such as cancer. A large body of evidence indicates that the dysregulation of some PKMTs leads to tumorigenesis via their non-histone substrates. However, most studies on other PKMTs have made slow progress owing to the lack of approaches for extensive screening of lysine methylation sites. However, recently, there has been a series of publications to perform large-scale analysis of protein lysine methylation. In this unit, we introduce a protocol for the global analysis of protein lysine methylation in cells by means of immunoaffinity enrichment and mass spectrometry. © 2016 by John Wiley & Sons, Inc.
Keywords: immunoaffinity enrichment; mass spectrometry; methylation; non-histone; post-translational modification; proteomics.
Copyright © 2016 John Wiley & Sons, Inc.
Figures
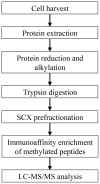

References
-
- Autiero M, Luttun A, Tjwa M, Carmeliet P. Placental growth factor and its receptor, vascular endothelial growth factor receptor-1: novel targets for stimulation of ischemic tissue revascularization and inhibition of angiogenic and inflammatory disorders. Journal of thrombosis and haemostasis : JTH. 2003;1:1356–1370. - PubMed
-
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nature biotechnology. 2006;24:1285–1292. - PubMed
-
- Bremang M, Cuomo A, Agresta AM, Stugiewicz M, Spadotto V, Bonaldi T. Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Molecular bioSystems. 2013;9:2231–2247. - PubMed
-
- Chi H, He K, Yang B, Chen Z, Sun RX, Fan SB, Zhang K, Liu C, Yuan ZF, Wang QH, Liu SQ, Dong MQ, He SM. pFind-Alioth: A novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. Journal of proteomics. 2015;125:89–97. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

