Translocation as continuous movement through the ribosome
- PMID: 27801619
- PMCID: PMC5207377
- DOI: 10.1080/15476286.2016.1240140
Translocation as continuous movement through the ribosome
Abstract
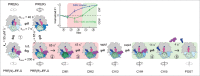
In each round of translation elongation, tRNAs and mRNA move within the ribosome by one codon at a time. tRNA-mRNA translocation is promoted by elongation factor G (EF-G) at the cost of GTP hydrolysis. The key questions for understanding translocation are how and when the tRNAs move and how EF-G coordinates motions of the ribosomal subunits with tRNA movement. Here we present 2 recent papers which describe the choreography of movements over the whole trajectory of translocation. We present the view that EF-G accelerates translocation by promoting the steps that lead to GTPase-dependent ribosome unlocking. EF-G facilitates the formation of the rotated state of the ribosome and uncouples the backward motions of the ribosomal subunits, forming an open conformation in which the tRNAs can rapidly move. Ribosome dynamics are important not only in translocation, but also in recoding events, such as frameshifting and bypassing, and mediate sensitivity to antibiotics.
Keywords: EF-G; mRNA; molecular machines; protein synthesis; ribosome; tRNA; translation; translation elongation; translocation.
Figures

References
-
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 2000; 406:318-22; PMID:10917535; http://dx.doi.org/10.1038/35018597 - DOI - PubMed
-
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature 1989; 342:142-8; PMID:2682263; http://dx.doi.org/10.1038/342142a0 - DOI - PubMed
-
- Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci U S A 2013; 110:20994-9; PMID:24324137; http://dx.doi.org/10.1073/pnas.1311423110 - DOI - PMC - PubMed
-
- Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science 2013; 340:1235970; PMID:23812721; http://dx.doi.org/10.1126/science.1235970 - DOI - PMC - PubMed
-
- Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 2013; 340:1236086; PMID:23812722; http://dx.doi.org/10.1126/science.1236086 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
