Genome of 'Ca. Desulfovibrio trichonymphae', an H2-oxidizing bacterium in a tripartite symbiotic system within a protist cell in the termite gut
- PMID: 27801909
- PMCID: PMC5322295
- DOI: 10.1038/ismej.2016.143
Genome of 'Ca. Desulfovibrio trichonymphae', an H2-oxidizing bacterium in a tripartite symbiotic system within a protist cell in the termite gut
Abstract
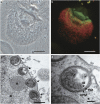
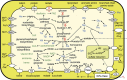
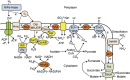
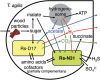
The cellulolytic protist Trichonympha agilis in the termite gut permanently hosts two symbiotic bacteria, 'Candidatus Endomicrobium trichonymphae' and 'Candidatus Desulfovibrio trichonymphae'. The former is an intracellular symbiont, and the latter is almost intracellular but still connected to the outside via a small pore. The complete genome of 'Ca. Endomicrobium trichonymphae' has previously been reported, and we here present the complete genome of 'Ca. Desulfovibrio trichonymphae'. The genome is small (1 410 056 bp), has many pseudogenes, and retains biosynthetic pathways for various amino acids and cofactors, which are partially complementary to those of 'Ca. Endomicrobium trichonymphae'. An amino acid permease gene has apparently been transferred between the ancestors of these two symbionts; a lateral gene transfer has affected their metabolic capacity. Notably, 'Ca. Desulfovibrio trichonymphae' retains the complex system to oxidize hydrogen by sulfate and/or fumarate, while genes for utilizing other substrates common in desulfovibrios are pseudogenized or missing. Thus, 'Ca. Desulfovibrio trichonymphae' is specialized to consume hydrogen that may otherwise inhibit fermentation processes in both T. agilis and 'Ca. Endomicrobium trichonymphae'. The small pore may be necessary to take up sulfate. This study depicts a genome-based model of a multipartite symbiotic system within a cellulolytic protist cell in the termite gut.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Badziong W, Ditter B, Thauer RK. (1979). Acetate and carbon dioxide assimilation by Desulfovibrio vulgaris (Marburg), growing on hydrogen and sulfate as sole energy source. Arch Microbiol 123: 301–305.
-
- Brune A. (2014). Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12: 168–180. - PubMed
-
- Cypionka H. (2000). Oxygen respiration by Desulfovibrio species. Annu Rev Microbiol 54: 827–848. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

