Transient ectopic expression of the histone demethylase JMJD3 accelerates the differentiation of human pluripotent stem cells
- PMID: 27802135
- PMCID: PMC5087640
- DOI: 10.1242/dev.139360
Transient ectopic expression of the histone demethylase JMJD3 accelerates the differentiation of human pluripotent stem cells
Abstract
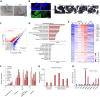
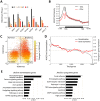
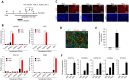
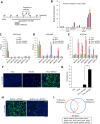
Harnessing epigenetic regulation is crucial for the efficient and proper differentiation of pluripotent stem cells (PSCs) into desired cell types. Histone H3 lysine 27 trimethylation (H3K27me3) functions as a barrier against cell differentiation through the suppression of developmental gene expression in PSCs. Here, we have generated human PSC (hPSC) lines in which genome-wide reduction of H3K27me3 can be induced by ectopic expression of the catalytic domain of the histone demethylase JMJD3 (called JMJD3c). We found that transient, forced demethylation of H3K27me3 alone triggers the upregulation of mesoendodermal genes, even when the culture conditions for the hPSCs are not changed. Furthermore, transient and forced expression of JMJD3c followed by the forced expression of lineage-defining transcription factors enabled the hPSCs to activate tissue-specific genes directly. We have also shown that the introduction of JMJD3c facilitates the differentiation of hPSCs into functional hepatic cells and skeletal muscle cells. These results suggest the utility of the direct manipulation of epigenomes for generating desired cell types from hPSCs for cell transplantation therapy and platforms for drug screenings.
Keywords: Hepatocytes; Histone demethylase; Human pluripotent stem cells; JMJD3; KDM6B; Skeletal muscle.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures

References
-
- Akiyama T., Xin L., Oda M., Sharov A. A., Amano M., Piao Y., Cadet J. S., Dudekula D. B., Qian Y., Wang W. et al. (2015). Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 22, 307-318. 10.1093/dnares/dsv013 - DOI - PMC - PubMed
-
- Akutsu H., Machida M., Kanzaki S., Sugawara T., Ohkura T., Nakamura N., Yamazaki-Inoue M., Miura T., Vemuri M., Rao M. S. et al. (2015). Xenogeneic-free defined conditions for derivation and expansion of human embryonic stem cells with mesenchymal stem cells. Regen. Ther. 1, 18-29. 10.1016/j.reth.2014.12.004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

